Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3790-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3790-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 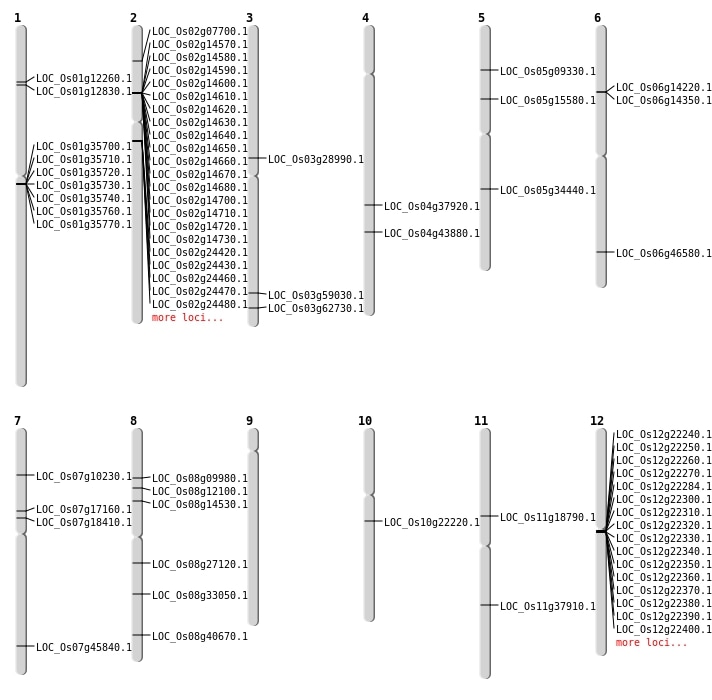
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10037269 | G-A | HET | ||
| SBS | Chr1 | 29913921 | G-A | HET | ||
| SBS | Chr10 | 11497152 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g22220 |
| SBS | Chr10 | 459585 | C-G | HET | ||
| SBS | Chr10 | 878262 | T-A | HET | ||
| SBS | Chr11 | 10667584 | G-A | Homo | STOP_GAINED | LOC_Os11g18790 |
| SBS | Chr12 | 8150281 | A-G | HET | ||
| SBS | Chr2 | 12541322 | C-G | HET | ||
| SBS | Chr2 | 14640203 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g25170 |
| SBS | Chr2 | 2086172 | C-T | HET | ||
| SBS | Chr2 | 22354305 | C-A | HET | ||
| SBS | Chr2 | 22463935 | T-A | HET | ||
| SBS | Chr2 | 28038436 | C-T | HET | ||
| SBS | Chr2 | 4119779 | G-A | HET | ||
| SBS | Chr3 | 11930189 | G-A | HET | ||
| SBS | Chr3 | 1255138 | A-T | HET | ||
| SBS | Chr3 | 16457725 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g28990 |
| SBS | Chr3 | 1750144 | C-T | HET | ||
| SBS | Chr3 | 19696004 | C-T | HET | ||
| SBS | Chr3 | 22165228 | A-C | HET | ||
| SBS | Chr3 | 26428418 | G-A | HET | ||
| SBS | Chr3 | 30294910 | G-A | Homo | ||
| SBS | Chr4 | 11172300 | C-A | Homo | ||
| SBS | Chr4 | 30454485 | G-T | Homo | ||
| SBS | Chr4 | 8890454 | C-T | HET | ||
| SBS | Chr5 | 24897450 | G-C | HET | ||
| SBS | Chr5 | 5075546 | T-C | HET | ||
| SBS | Chr5 | 5902858 | T-G | HET | ||
| SBS | Chr6 | 10787475 | T-A | HET | ||
| SBS | Chr6 | 21820800 | C-G | HET | ||
| SBS | Chr6 | 25653899 | C-T | HET | ||
| SBS | Chr6 | 5998268 | A-G | HET | ||
| SBS | Chr7 | 18371356 | G-A | HET | ||
| SBS | Chr7 | 27358506 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g45840 |
| SBS | Chr8 | 10879557 | A-G | Homo | ||
| SBS | Chr8 | 21260309 | T-C | Homo | ||
| SBS | Chr8 | 22420803 | A-C | Homo | ||
| SBS | Chr8 | 25752811 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g40670 |
| SBS | Chr8 | 27876488 | A-G | Homo | ||
| SBS | Chr8 | 27937548 | G-A | Homo | ||
| SBS | Chr8 | 3474122 | G-A | Homo |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 2417095 | 2417099 | 4 | |
| Deletion | Chr11 | 2536556 | 2536559 | 3 | |
| Deletion | Chr2 | 4015220 | 4015230 | 10 | LOC_Os02g07700 |
| Deletion | Chr2 | 8034001 | 8148000 | 113999 | 16 |
| Deletion | Chr9 | 8847438 | 8847444 | 6 | |
| Deletion | Chr2 | 9114994 | 9114996 | 2 | |
| Deletion | Chr7 | 10082001 | 10091000 | 8999 | LOC_Os07g17160 |
| Deletion | Chr7 | 10900784 | 10900793 | 9 | LOC_Os07g18410 |
| Deletion | Chr2 | 11613500 | 11613501 | 1 | |
| Deletion | Chr11 | 12087648 | 12087651 | 3 | |
| Deletion | Chr8 | 12094810 | 12094819 | 9 | |
| Deletion | Chr12 | 12534001 | 12857000 | 322999 | 53 |
| Deletion | Chr3 | 12821911 | 12821916 | 5 | |
| Deletion | Chr12 | 12901001 | 13051000 | 149999 | 18 |
| Deletion | Chr12 | 13054001 | 13105000 | 50999 | 10 |
| Deletion | Chr12 | 13661484 | 13661485 | 1 | |
| Deletion | Chr2 | 14155001 | 14236000 | 80999 | 12 |
| Deletion | Chr1 | 14334691 | 14334700 | 9 | |
| Deletion | Chr2 | 16321392 | 16321395 | 3 | LOC_Os02g27560 |
| Deletion | Chr9 | 18606356 | 18606364 | 8 | |
| Deletion | Chr1 | 19697210 | 19697219 | 9 | |
| Deletion | Chr1 | 19767001 | 19793000 | 25999 | 4 |
| Deletion | Chr1 | 19800001 | 19817000 | 16999 | 3 |
| Deletion | Chr5 | 22186739 | 22186746 | 7 | |
| Deletion | Chr12 | 24011544 | 24011548 | 4 | |
| Deletion | Chr1 | 28601773 | 28601777 | 4 | |
| Deletion | Chr3 | 29954923 | 29954925 | 2 | |
| Deletion | Chr3 | 35499984 | 35499991 | 7 | LOC_Os03g62730 |
Insertions: 7
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 6702839 | 7100505 | 2 |
| Inversion | Chr1 | 6702869 | 6805194 | LOC_Os01g12260 |
| Inversion | Chr12 | 12853925 | 13101827 | 2 |
| Inversion | Chr12 | 12900681 | 13267469 | LOC_Os12g23450 |
| Inversion | Chr11 | 22488848 | 22490278 | LOC_Os11g37910 |
Translocations: 17
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 1224682 | Chr4 | 27248223 | |
| Translocation | Chr9 | 5002830 | Chr3 | 2456147 | |
| Translocation | Chr10 | 5039498 | Chr8 | 16573989 | LOC_Os08g27120 |
| Translocation | Chr7 | 5492724 | Chr3 | 33599334 | 2 |
| Translocation | Chr8 | 5771681 | Chr6 | 8004069 | 2 |
| Translocation | Chr8 | 7125835 | Chr5 | 8822401 | 2 |
| Translocation | Chr8 | 8734095 | Chr2 | 23342582 | 2 |
| Translocation | Chr12 | 13101813 | Chr6 | 7934013 | 2 |
| Translocation | Chr8 | 13598939 | Chr3 | 16448936 | |
| Translocation | Chr8 | 13598943 | Chr3 | 16448819 | |
| Translocation | Chr9 | 14647029 | Chr6 | 28280471 | LOC_Os06g46580 |
| Translocation | Chr8 | 14932717 | Chr5 | 20403560 | LOC_Os05g34440 |
| Translocation | Chr7 | 16998626 | Chr4 | 22551334 | LOC_Os04g37920 |
| Translocation | Chr8 | 20533835 | Chr4 | 7877662 | LOC_Os08g33050 |
| Translocation | Chr9 | 20826319 | Chr3 | 17136842 | |
| Translocation | Chr6 | 23516344 | Chr5 | 5212207 | LOC_Os05g09330 |
| Translocation | Chr4 | 25990483 | Chr2 | 26794030 | LOC_Os04g43880 |
