Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3792-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3792-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 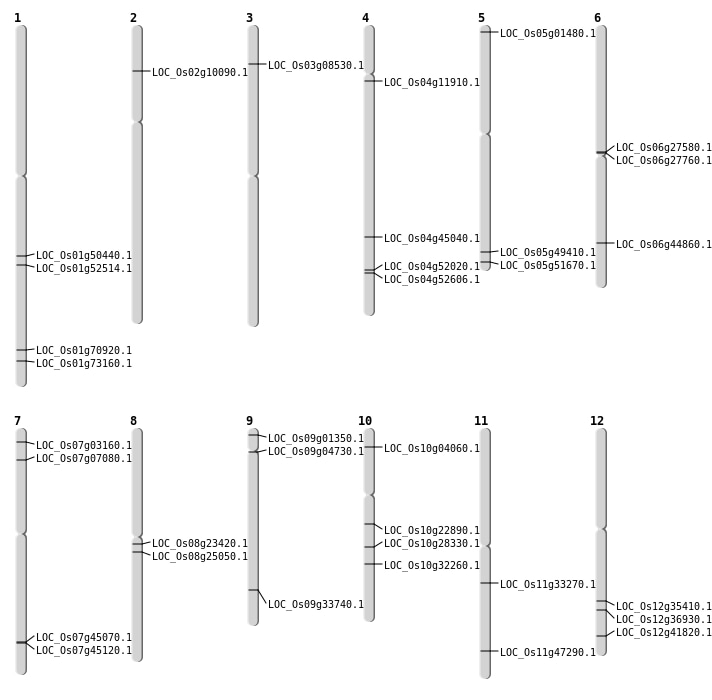
|
| Variant Information |
|---|
Single base substitutions: 23
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 25465418 | A-T | HET | ||
| SBS | Chr1 | 39759385 | C-T | Homo | ||
| SBS | Chr10 | 3706593 | A-T | HET | ||
| SBS | Chr11 | 7391556 | G-T | HET | ||
| SBS | Chr11 | 7887030 | T-G | HET | ||
| SBS | Chr12 | 10844427 | G-A | Homo | ||
| SBS | Chr12 | 14273904 | T-C | Homo | ||
| SBS | Chr12 | 1523116 | A-G | HET | ||
| SBS | Chr12 | 21090737 | C-G | Homo | ||
| SBS | Chr2 | 25428922 | C-T | HET | ||
| SBS | Chr2 | 5692311 | A-G | Homo | ||
| SBS | Chr3 | 11933649 | G-A | HET | ||
| SBS | Chr3 | 19112117 | G-T | HET | ||
| SBS | Chr3 | 22829807 | G-T | HET | ||
| SBS | Chr3 | 26932850 | C-A | HET | ||
| SBS | Chr4 | 15290429 | T-C | Homo | ||
| SBS | Chr4 | 22584338 | C-T | HET | ||
| SBS | Chr4 | 26826213 | A-G | HET | ||
| SBS | Chr5 | 7292995 | A-G | HET | ||
| SBS | Chr6 | 15610684 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g27580 |
| SBS | Chr7 | 20264264 | G-A | HET | ||
| SBS | Chr7 | 3494647 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g07080 |
| SBS | Chr9 | 14757858 | C-T | Homo |
Deletions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 882520 | 882524 | 4 | |
| Deletion | Chr5 | 2129913 | 2129920 | 7 | |
| Deletion | Chr4 | 6532946 | 6532953 | 7 | LOC_Os04g11910 |
| Deletion | Chr10 | 11896710 | 11896711 | 1 | LOC_Os10g22890 |
| Deletion | Chr12 | 11911824 | 11911825 | 1 | |
| Deletion | Chr12 | 17945996 | 17946001 | 5 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr5 | 12758792 | 12758792 | 1 |
Inversions: 44
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 279756 | 279865 | LOC_Os09g01350 |
| Inversion | Chr5 | 286292 | 286584 | LOC_Os05g01480 |
| Inversion | Chr6 | 592724 | 593156 | |
| Inversion | Chr4 | 1042267 | 1042425 | |
| Inversion | Chr7 | 1237722 | 1237930 | LOC_Os07g03160 |
| Inversion | Chr10 | 1872462 | 1872600 | LOC_Os10g04060 |
| Inversion | Chr10 | 2254455 | 2254751 | |
| Inversion | Chr9 | 2514975 | 2515221 | LOC_Os09g04730 |
| Inversion | Chr11 | 3900150 | 3900412 | |
| Inversion | Chr3 | 4384454 | 4384742 | LOC_Os03g08530 |
| Inversion | Chr2 | 5255141 | 5255425 | LOC_Os02g10090 |
| Inversion | Chr5 | 5748201 | 5748506 | |
| Inversion | Chr9 | 6542126 | 6542327 | |
| Inversion | Chr7 | 7343189 | 7343333 | |
| Inversion | Chr5 | 7575254 | 7575510 | |
| Inversion | Chr2 | 8896375 | 8896552 | |
| Inversion | Chr1 | 9625688 | 9626220 | |
| Inversion | Chr9 | 12816353 | 12816649 | |
| Inversion | Chr6 | 13319527 | 13319747 | |
| Inversion | Chr4 | 13393116 | 13393376 | |
| Inversion | Chr8 | 14166230 | 14166424 | LOC_Os08g23420 |
| Inversion | Chr10 | 14741339 | 14741693 | LOC_Os10g28330 |
| Inversion | Chr8 | 15200908 | 15201025 | LOC_Os08g25050 |
| Inversion | Chr10 | 16935510 | 16935777 | LOC_Os10g32260 |
| Inversion | Chr11 | 19677805 | 19678349 | LOC_Os11g33270 |
| Inversion | Chr5 | 19759958 | 19760278 | |
| Inversion | Chr12 | 21533182 | 21533498 | LOC_Os12g35410 |
| Inversion | Chr9 | 21795125 | 21795335 | |
| Inversion | Chr12 | 22642006 | 22642285 | LOC_Os12g36930 |
| Inversion | Chr3 | 24559334 | 24559618 | |
| Inversion | Chr12 | 25902304 | 25902594 | LOC_Os12g41820 |
| Inversion | Chr7 | 26896273 | 26896613 | LOC_Os07g45070 |
| Inversion | Chr7 | 26936054 | 26936319 | LOC_Os07g45120 |
| Inversion | Chr6 | 27117142 | 27117339 | LOC_Os06g44860 |
| Inversion | Chr11 | 27316524 | 27316709 | |
| Inversion | Chr5 | 28330503 | 28330608 | LOC_Os05g49410 |
| Inversion | Chr1 | 28957019 | 28957154 | LOC_Os01g50440 |
| Inversion | Chr5 | 29634832 | 29635133 | LOC_Os05g51670 |
| Inversion | Chr1 | 30178445 | 30178603 | LOC_Os01g52514 |
| Inversion | Chr4 | 30892955 | 30893205 | LOC_Os04g52020 |
| Inversion | Chr4 | 31287619 | 31287863 | LOC_Os04g52606 |
| Inversion | Chr1 | 38953242 | 38953357 | |
| Inversion | Chr1 | 41053263 | 41053448 | LOC_Os01g70920 |
| Inversion | Chr1 | 42426793 | 42427117 | LOC_Os01g73160 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 13456017 | Chr4 | 9486025 | |
| Translocation | Chr6 | 16040202 | Chr4 | 11855994 | |
| Translocation | Chr4 | 17771619 | Chr2 | 19422122 | |
| Translocation | Chr12 | 19030375 | Chr11 | 28433553 | LOC_Os11g47290 |
| Translocation | Chr9 | 19929206 | Chr4 | 26648747 | 2 |
| Translocation | Chr4 | 23938474 | Chr3 | 5772805 | |
| Translocation | Chr12 | 24881478 | Chr6 | 15720160 | LOC_Os06g27760 |
| Translocation | Chr4 | 32897644 | Chr1 | 39114202 |
