Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3815-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3815-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 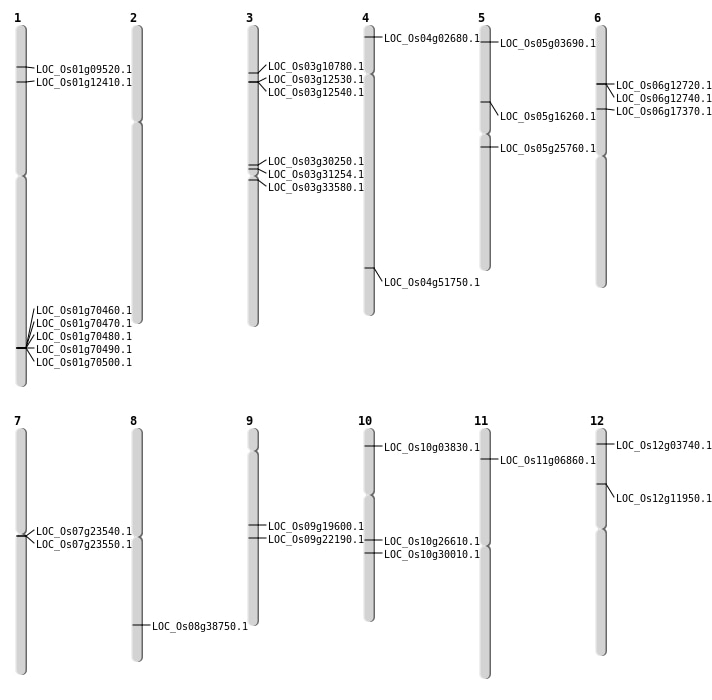
|
| Variant Information |
|---|
Single base substitutions: 93
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17034766 | G-A | HET | ||
| SBS | Chr1 | 20297544 | T-A | HET | ||
| SBS | Chr1 | 21181309 | G-A | HET | ||
| SBS | Chr1 | 24425056 | A-T | HET | ||
| SBS | Chr1 | 24860678 | C-A | HET | ||
| SBS | Chr1 | 26914163 | A-G | Homo | ||
| SBS | Chr1 | 26914168 | C-T | Homo | ||
| SBS | Chr1 | 33673121 | G-T | HET | ||
| SBS | Chr1 | 3503732 | C-T | HET | ||
| SBS | Chr1 | 3503750 | A-T | HET | ||
| SBS | Chr1 | 35798502 | C-A | HET | ||
| SBS | Chr1 | 37940721 | C-A | HET | ||
| SBS | Chr1 | 39192244 | G-T | HET | ||
| SBS | Chr1 | 6783260 | G-T | Homo | STOP_GAINED | LOC_Os01g12410 |
| SBS | Chr1 | 7032897 | G-T | HET | ||
| SBS | Chr1 | 9993712 | G-A | HET | ||
| SBS | Chr10 | 13874469 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g26610 |
| SBS | Chr10 | 15175618 | G-A | HET | ||
| SBS | Chr10 | 15576278 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g30010 |
| SBS | Chr10 | 15576279 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g30010 |
| SBS | Chr10 | 1738415 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g03830 |
| SBS | Chr10 | 20309959 | G-T | HET | ||
| SBS | Chr10 | 3523712 | G-C | HET | ||
| SBS | Chr10 | 830542 | T-A | HET | ||
| SBS | Chr11 | 11613509 | G-T | HET | ||
| SBS | Chr11 | 20071011 | G-T | HET | ||
| SBS | Chr11 | 3367899 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g06860 |
| SBS | Chr11 | 5830298 | G-T | HET | ||
| SBS | Chr11 | 9674140 | G-A | Homo | ||
| SBS | Chr12 | 14100564 | G-T | HET | ||
| SBS | Chr12 | 1515065 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g03740 |
| SBS | Chr12 | 16312683 | A-T | HET | ||
| SBS | Chr12 | 17227008 | A-G | HET | ||
| SBS | Chr12 | 17419465 | G-T | HET | ||
| SBS | Chr12 | 2459523 | G-T | HET | ||
| SBS | Chr12 | 601623 | T-A | HET | ||
| SBS | Chr12 | 6526181 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g11950 |
| SBS | Chr2 | 16123901 | A-C | HET | ||
| SBS | Chr2 | 25066375 | G-A | HET | ||
| SBS | Chr2 | 25490056 | G-A | HET | ||
| SBS | Chr2 | 27867175 | C-T | HET | ||
| SBS | Chr2 | 28776982 | C-T | HET | ||
| SBS | Chr2 | 29842139 | A-T | HET | ||
| SBS | Chr2 | 30051249 | C-A | HET | ||
| SBS | Chr3 | 15375096 | G-T | HET | ||
| SBS | Chr3 | 16251751 | A-T | HET | ||
| SBS | Chr3 | 17261313 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g30250 |
| SBS | Chr3 | 19776116 | A-T | HET | ||
| SBS | Chr3 | 19776117 | G-T | HET | ||
| SBS | Chr3 | 20944062 | C-A | HET | ||
| SBS | Chr3 | 21100983 | G-T | HET | ||
| SBS | Chr3 | 26786767 | T-A | HET | ||
| SBS | Chr3 | 8622425 | A-T | HET | ||
| SBS | Chr3 | 9897008 | A-G | HET | ||
| SBS | Chr4 | 1018068 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g02680 |
| SBS | Chr4 | 11248828 | G-C | HET | ||
| SBS | Chr4 | 11296381 | A-G | HET | ||
| SBS | Chr4 | 2375453 | A-G | HET | ||
| SBS | Chr4 | 31931227 | G-T | HET | ||
| SBS | Chr4 | 5727381 | G-C | HET | ||
| SBS | Chr5 | 10609328 | T-A | HET | ||
| SBS | Chr5 | 14988302 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g25760 |
| SBS | Chr5 | 1605692 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g03690 |
| SBS | Chr5 | 22532982 | T-A | HET | ||
| SBS | Chr5 | 28263398 | T-A | HET | ||
| SBS | Chr5 | 29592525 | C-A | HET | ||
| SBS | Chr5 | 9205643 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g16260 |
| SBS | Chr5 | 9323130 | C-A | HET | ||
| SBS | Chr6 | 10066118 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g17370 |
| SBS | Chr6 | 23060694 | A-G | HET | ||
| SBS | Chr6 | 24077646 | C-T | HET | ||
| SBS | Chr6 | 244413 | T-A | HET | ||
| SBS | Chr6 | 6947743 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g12720 |
| SBS | Chr6 | 6954226 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g12740 |
| SBS | Chr6 | 824207 | C-A | HET | ||
| SBS | Chr7 | 13297476 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g23540 |
| SBS | Chr7 | 13297480 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g23540 |
| SBS | Chr7 | 13314738 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g23550 |
| SBS | Chr7 | 13907248 | G-A | HET | ||
| SBS | Chr7 | 25561729 | C-A | HET | ||
| SBS | Chr7 | 26101881 | G-A | HET | ||
| SBS | Chr7 | 26909102 | G-A | Homo | ||
| SBS | Chr8 | 12878585 | T-A | HET | ||
| SBS | Chr8 | 17835621 | G-C | HET | ||
| SBS | Chr8 | 18628528 | T-A | HET | ||
| SBS | Chr8 | 20990634 | C-A | HET | ||
| SBS | Chr8 | 24500628 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g38750 |
| SBS | Chr8 | 25303922 | A-T | HET | ||
| SBS | Chr9 | 10866806 | G-A | HET | ||
| SBS | Chr9 | 11720553 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g19600 |
| SBS | Chr9 | 17454713 | A-T | HET | ||
| SBS | Chr9 | 17727876 | G-T | HET | ||
| SBS | Chr9 | 21670529 | G-T | HET |
Deletions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 3029982 | 3029984 | 2 | |
| Deletion | Chr12 | 3381555 | 3381583 | 28 | |
| Deletion | Chr3 | 6634001 | 6647000 | 12999 | 2 |
| Deletion | Chr4 | 11247491 | 11247495 | 4 | |
| Deletion | Chr7 | 15677116 | 15677122 | 6 | |
| Deletion | Chr3 | 17802504 | 17802506 | 2 | LOC_Os03g31254 |
| Deletion | Chr1 | 22181176 | 22181177 | 1 | |
| Deletion | Chr5 | 22287068 | 22287069 | 1 | |
| Deletion | Chr1 | 32133361 | 32133362 | 1 | |
| Deletion | Chr3 | 34244772 | 34244776 | 4 | |
| Deletion | Chr1 | 40805001 | 40836000 | 30999 | 5 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 2805544 | 2805544 | 1 | |
| Insertion | Chr12 | 17878258 | 17878258 | 1 | |
| Insertion | Chr3 | 19184302 | 19184303 | 2 | LOC_Os03g33580 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 5511672 | 5545127 | LOC_Os03g10780 |
| Inversion | Chr3 | 5511723 | 5545153 | LOC_Os03g10780 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 18984728 | Chr2 | 3226135 | |
| Translocation | Chr11 | 28270432 | Chr9 | 13425257 | LOC_Os09g22190 |
| Translocation | Chr4 | 30670307 | Chr1 | 4858483 | 2 |
