Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3830-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3830-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 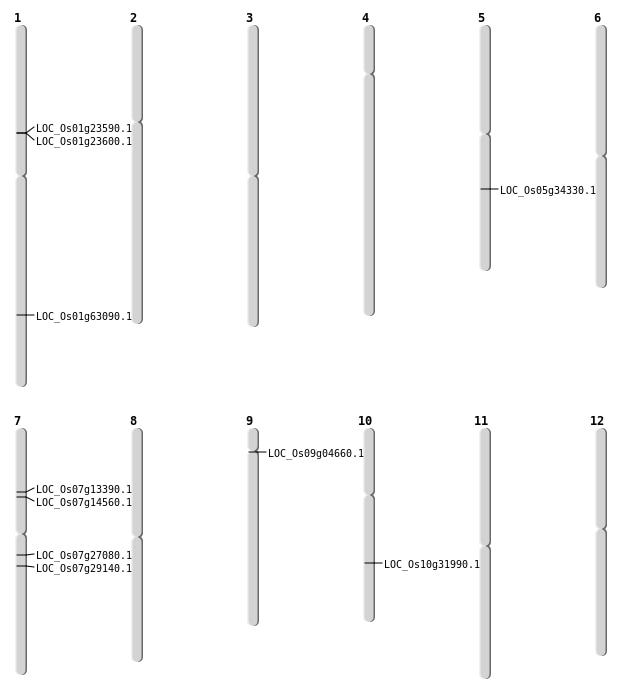
|
| Variant Information |
|---|
Single base substitutions: 50
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20506128 | C-T | HET | ||
| SBS | Chr1 | 24489998 | G-A | HET | ||
| SBS | Chr1 | 36562249 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g63090 |
| SBS | Chr1 | 36695200 | G-T | HET | ||
| SBS | Chr1 | 38631803 | G-A | HET | ||
| SBS | Chr10 | 15103539 | T-A | HET | ||
| SBS | Chr10 | 16641526 | A-G | HET | ||
| SBS | Chr10 | 16773683 | C-T | HET | ||
| SBS | Chr10 | 227294 | G-A | HET | ||
| SBS | Chr11 | 17793593 | A-C | HET | ||
| SBS | Chr11 | 24648923 | A-G | Homo | ||
| SBS | Chr11 | 7589979 | C-G | HET | ||
| SBS | Chr12 | 1375929 | C-T | HET | ||
| SBS | Chr12 | 17057241 | C-T | HET | ||
| SBS | Chr12 | 17793899 | G-A | HET | ||
| SBS | Chr12 | 21475791 | T-G | HET | ||
| SBS | Chr12 | 7482786 | G-A | HET | ||
| SBS | Chr12 | 8434952 | T-C | HET | ||
| SBS | Chr2 | 16720148 | G-A | HET | ||
| SBS | Chr2 | 17798782 | C-T | HET | ||
| SBS | Chr2 | 294985 | A-T | Homo | ||
| SBS | Chr2 | 32999160 | A-C | HET | ||
| SBS | Chr2 | 5965931 | C-T | HET | ||
| SBS | Chr2 | 8017356 | G-A | HET | ||
| SBS | Chr3 | 11782205 | G-C | HET | ||
| SBS | Chr3 | 16430760 | T-C | HET | ||
| SBS | Chr4 | 12489794 | C-T | HET | ||
| SBS | Chr4 | 15491741 | G-A | HET | ||
| SBS | Chr4 | 25720165 | G-T | HET | ||
| SBS | Chr4 | 34520290 | T-A | Homo | ||
| SBS | Chr4 | 9483807 | T-C | HET | ||
| SBS | Chr5 | 17749882 | T-A | HET | ||
| SBS | Chr6 | 15903697 | T-C | Homo | ||
| SBS | Chr6 | 17902185 | G-A | HET | ||
| SBS | Chr6 | 22007714 | C-A | HET | ||
| SBS | Chr7 | 15693820 | A-G | HET | ||
| SBS | Chr7 | 15694786 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g27080 |
| SBS | Chr7 | 17065022 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g29140 |
| SBS | Chr7 | 26333348 | T-C | HET | ||
| SBS | Chr7 | 5490354 | C-T | Homo | ||
| SBS | Chr8 | 13162145 | A-T | HET | ||
| SBS | Chr8 | 17032819 | T-C | HET | ||
| SBS | Chr8 | 18606162 | G-A | HET | ||
| SBS | Chr8 | 21294800 | T-A | HET | ||
| SBS | Chr8 | 22309112 | C-T | HET | ||
| SBS | Chr8 | 5036482 | C-A | HET | ||
| SBS | Chr8 | 9008589 | C-T | HET | ||
| SBS | Chr9 | 18651616 | G-A | HET | ||
| SBS | Chr9 | 2485929 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g04660 |
| SBS | Chr9 | 4495479 | T-A | HET |
Deletions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 662293 | 662295 | 2 | |
| Deletion | Chr5 | 4380150 | 4380152 | 2 | |
| Deletion | Chr11 | 4407213 | 4407216 | 3 | |
| Deletion | Chr4 | 4723848 | 4723855 | 7 | |
| Deletion | Chr3 | 9615539 | 9615545 | 6 | |
| Deletion | Chr6 | 11668089 | 11668091 | 2 | |
| Deletion | Chr1 | 13252001 | 13267000 | 14999 | 2 |
| Deletion | Chr9 | 14199624 | 14199633 | 9 | |
| Deletion | Chr2 | 14239764 | 14239766 | 2 | |
| Deletion | Chr6 | 14794499 | 14794500 | 1 | |
| Deletion | Chr12 | 14961329 | 14961348 | 19 | |
| Deletion | Chr10 | 16808645 | 16808646 | 1 | LOC_Os10g31990 |
| Deletion | Chr8 | 23088125 | 23088133 | 8 | |
| Deletion | Chr2 | 32225960 | 32225962 | 2 | |
| Deletion | Chr1 | 34128643 | 34128645 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 6884165 | 6884166 | 2 | |
| Insertion | Chr9 | 21027694 | 21027695 | 2 | |
| Insertion | Chr9 | 7314134 | 7314135 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 7676399 | 8306180 | 2 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 15900307 | Chr6 | 25107112 | |
| Translocation | Chr11 | 17270060 | Chr5 | 20336850 | LOC_Os05g34330 |
