Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3834-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3834-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 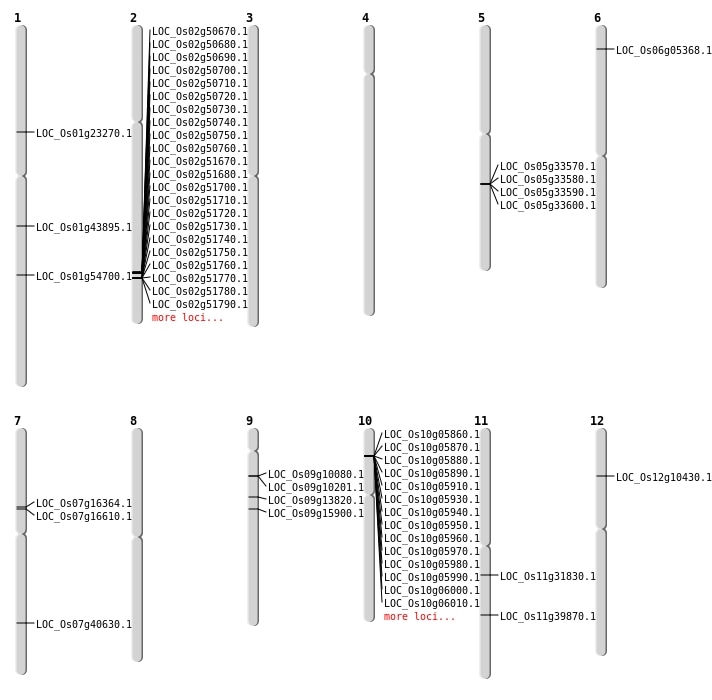
|
| Variant Information |
|---|
Single base substitutions: 59
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13077455 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g23270 |
| SBS | Chr1 | 19787204 | G-A | HET | ||
| SBS | Chr1 | 20335370 | C-T | HET | ||
| SBS | Chr1 | 27423195 | C-T | HET | ||
| SBS | Chr1 | 31476191 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g54700 |
| SBS | Chr1 | 35400654 | A-G | HET | ||
| SBS | Chr1 | 39813437 | G-A | HET | ||
| SBS | Chr1 | 7156200 | T-A | Homo | ||
| SBS | Chr1 | 7710722 | G-A | Homo | ||
| SBS | Chr10 | 20017472 | C-T | HET | ||
| SBS | Chr10 | 9537711 | G-A | HET | ||
| SBS | Chr11 | 11678473 | C-T | Homo | ||
| SBS | Chr11 | 15173681 | T-A | Homo | ||
| SBS | Chr11 | 18707645 | T-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g31830 |
| SBS | Chr11 | 20064422 | C-T | Homo | ||
| SBS | Chr11 | 23768054 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g39870 |
| SBS | Chr11 | 5435663 | A-T | Homo | ||
| SBS | Chr11 | 9235390 | C-T | HET | ||
| SBS | Chr12 | 10271759 | G-T | Homo | ||
| SBS | Chr12 | 14040481 | A-G | Homo | ||
| SBS | Chr12 | 22095033 | G-T | HET | ||
| SBS | Chr12 | 8847104 | C-A | HET | ||
| SBS | Chr2 | 32764606 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g53550 |
| SBS | Chr2 | 32863623 | C-T | HET | ||
| SBS | Chr3 | 10785461 | G-T | HET | ||
| SBS | Chr3 | 15601659 | A-T | HET | ||
| SBS | Chr3 | 15601663 | C-A | HET | ||
| SBS | Chr3 | 23687987 | G-A | HET | ||
| SBS | Chr3 | 2512140 | T-C | HET | ||
| SBS | Chr3 | 26845685 | A-G | HET | ||
| SBS | Chr3 | 26845687 | C-T | HET | ||
| SBS | Chr4 | 13810359 | C-T | HET | ||
| SBS | Chr4 | 17380560 | T-G | Homo | ||
| SBS | Chr4 | 17473067 | A-G | Homo | ||
| SBS | Chr4 | 22984498 | A-T | HET | ||
| SBS | Chr4 | 23955112 | T-C | HET | ||
| SBS | Chr4 | 3207302 | C-T | HET | ||
| SBS | Chr4 | 33376458 | C-T | HET | ||
| SBS | Chr5 | 12876176 | A-G | Homo | ||
| SBS | Chr5 | 18512760 | A-G | HET | ||
| SBS | Chr5 | 20214031 | C-T | HET | ||
| SBS | Chr5 | 27715090 | A-G | Homo | ||
| SBS | Chr5 | 5884877 | G-T | HET | ||
| SBS | Chr5 | 5884879 | G-T | HET | ||
| SBS | Chr6 | 16196322 | A-T | Homo | ||
| SBS | Chr6 | 2419806 | C-T | HET | STOP_GAINED | LOC_Os06g05368 |
| SBS | Chr6 | 4776556 | T-C | HET | ||
| SBS | Chr7 | 3156175 | G-A | HET | ||
| SBS | Chr7 | 9743586 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g16610 |
| SBS | Chr8 | 10974957 | T-C | HET | ||
| SBS | Chr8 | 22518414 | G-A | HET | ||
| SBS | Chr8 | 352229 | T-A | Homo | ||
| SBS | Chr9 | 12659697 | G-A | Homo | ||
| SBS | Chr9 | 16414276 | G-C | HET | ||
| SBS | Chr9 | 21840041 | T-C | HET | ||
| SBS | Chr9 | 2292374 | G-A | HET | ||
| SBS | Chr9 | 5495450 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g10080 |
| SBS | Chr9 | 8119236 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g13820 |
| SBS | Chr9 | 9715872 | T-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g15900 |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 2960001 | 2978000 | 17999 | 4 |
| Deletion | Chr10 | 2982001 | 3165000 | 182999 | 22 |
| Deletion | Chr10 | 3172001 | 3209000 | 36999 | 7 |
| Deletion | Chr10 | 3214001 | 3649000 | 434999 | 67 |
| Deletion | Chr8 | 4049363 | 4049368 | 5 | |
| Deletion | Chr12 | 5527186 | 5527207 | 21 | LOC_Os12g10430 |
| Deletion | Chr11 | 11614606 | 11614608 | 2 | |
| Deletion | Chr3 | 13272601 | 13272609 | 8 | |
| Deletion | Chr5 | 19733001 | 19759000 | 25999 | 4 |
| Deletion | Chr9 | 20377015 | 20377020 | 5 | |
| Deletion | Chr7 | 21066907 | 21066909 | 2 | |
| Deletion | Chr11 | 21267523 | 21267528 | 5 | |
| Deletion | Chr12 | 22564957 | 22564969 | 12 | |
| Deletion | Chr5 | 22625265 | 22625283 | 18 | |
| Deletion | Chr10 | 23077001 | 23101000 | 23999 | 7 |
| Deletion | Chr7 | 24349947 | 24349948 | 1 | LOC_Os07g40630 |
| Deletion | Chr2 | 30952001 | 30996000 | 43999 | 10 |
| Deletion | Chr2 | 31640001 | 31854000 | 213999 | 36 |
Insertions: 6
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 2195133 | 2954929 | |
| Inversion | Chr5 | 23923882 | 24026398 | |
| Inversion | Chr2 | 30951960 | 31147795 | |
| Inversion | Chr2 | 30997254 | 31147797 | LOC_Os02g50760 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 1589518 | Chr2 | 9833429 | |
| Translocation | Chr10 | 5484403 | Chr2 | 14658145 | |
| Translocation | Chr9 | 5556218 | Chr7 | 9565144 | 2 |
| Translocation | Chr12 | 11347786 | Chr4 | 15227358 | |
| Translocation | Chr6 | 17879908 | Chr1 | 25146875 | LOC_Os01g43895 |
| Translocation | Chr6 | 18236820 | Chr2 | 31864506 | |
| Translocation | Chr5 | 18985805 | Chr2 | 3226637 |
