Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3851-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3851-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 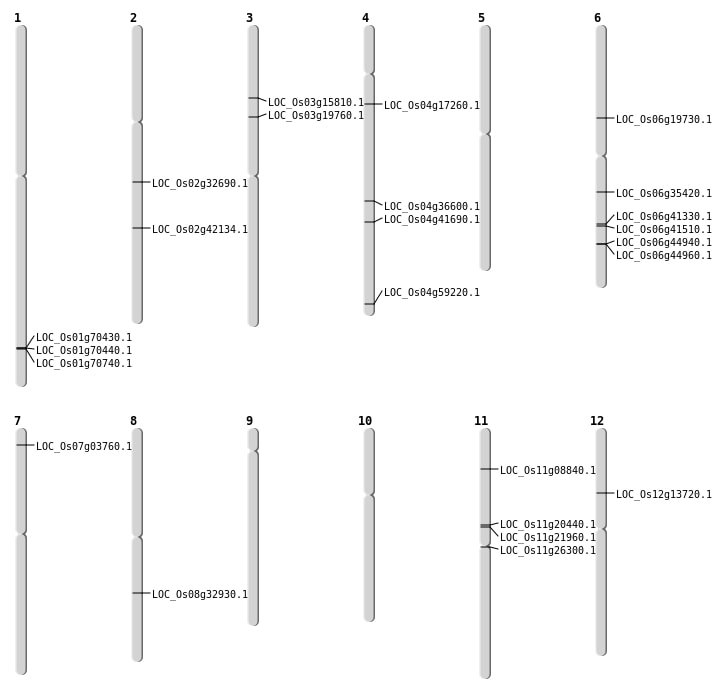
|
| Variant Information |
|---|
Single base substitutions: 69
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1024003 | T-C | HET | ||
| SBS | Chr1 | 1554791 | G-A | HET | ||
| SBS | Chr1 | 34112172 | G-A | Homo | ||
| SBS | Chr1 | 39945621 | C-T | HET | ||
| SBS | Chr10 | 10513723 | A-G | HET | ||
| SBS | Chr10 | 8639287 | G-A | HET | ||
| SBS | Chr11 | 11831256 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g20440 |
| SBS | Chr11 | 12875586 | A-C | HET | ||
| SBS | Chr11 | 13950708 | G-C | HET | ||
| SBS | Chr11 | 14318408 | C-G | HET | ||
| SBS | Chr11 | 15061182 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g26300 |
| SBS | Chr11 | 18758519 | G-A | HET | ||
| SBS | Chr11 | 3779519 | T-G | Homo | ||
| SBS | Chr11 | 482412 | A-G | HET | ||
| SBS | Chr12 | 13730765 | A-T | HET | ||
| SBS | Chr12 | 15661800 | T-C | HET | ||
| SBS | Chr12 | 2007052 | T-C | HET | ||
| SBS | Chr12 | 21616514 | C-A | HET | ||
| SBS | Chr12 | 26775070 | T-G | HET | ||
| SBS | Chr2 | 27594507 | C-T | HET | ||
| SBS | Chr2 | 30308369 | C-A | Homo | ||
| SBS | Chr2 | 8932331 | G-A | HET | ||
| SBS | Chr3 | 23981410 | A-G | HET | ||
| SBS | Chr3 | 32249801 | G-C | Homo | ||
| SBS | Chr3 | 33224948 | A-T | HET | ||
| SBS | Chr3 | 34047034 | C-T | Homo | ||
| SBS | Chr3 | 3759185 | C-A | HET | ||
| SBS | Chr3 | 4577625 | G-A | HET | ||
| SBS | Chr3 | 5460639 | C-A | HET | ||
| SBS | Chr3 | 7145805 | T-A | HET | ||
| SBS | Chr4 | 13692549 | C-T | HET | ||
| SBS | Chr4 | 22079078 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g36600 |
| SBS | Chr4 | 32801955 | T-C | HET | ||
| SBS | Chr4 | 35226393 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g59220 |
| SBS | Chr4 | 8183868 | G-C | HET | ||
| SBS | Chr4 | 9466966 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g17260 |
| SBS | Chr5 | 14088735 | G-A | Homo | ||
| SBS | Chr5 | 24432818 | G-A | HET | ||
| SBS | Chr5 | 24826360 | C-T | HET | ||
| SBS | Chr5 | 4756064 | C-T | HET | ||
| SBS | Chr5 | 7675888 | C-T | HET | ||
| SBS | Chr6 | 11553499 | A-T | HET | ||
| SBS | Chr6 | 24741348 | A-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g41330 |
| SBS | Chr6 | 24891009 | T-A | Homo | ||
| SBS | Chr6 | 24891018 | G-A | Homo | ||
| SBS | Chr6 | 24891019 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g41510 |
| SBS | Chr6 | 27179888 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g44940 |
| SBS | Chr6 | 4353714 | A-G | HET | ||
| SBS | Chr6 | 7214681 | A-G | Homo | ||
| SBS | Chr7 | 10917409 | T-C | Homo | ||
| SBS | Chr7 | 16924015 | A-C | Homo | ||
| SBS | Chr7 | 22935583 | C-A | HET | ||
| SBS | Chr7 | 24102170 | T-C | HET | ||
| SBS | Chr7 | 28385928 | G-C | HET | ||
| SBS | Chr7 | 28531362 | G-C | HET | ||
| SBS | Chr8 | 14264651 | A-T | HET | ||
| SBS | Chr8 | 17564089 | G-A | HET | ||
| SBS | Chr8 | 19029044 | T-C | HET | ||
| SBS | Chr8 | 22475860 | C-T | HET | ||
| SBS | Chr8 | 23339818 | A-G | HET | ||
| SBS | Chr8 | 26309951 | G-A | HET | ||
| SBS | Chr8 | 26309953 | C-A | HET | ||
| SBS | Chr9 | 10184006 | C-G | HET | ||
| SBS | Chr9 | 11641359 | C-T | HET | ||
| SBS | Chr9 | 16141038 | G-A | HET | ||
| SBS | Chr9 | 16781779 | G-A | HET | ||
| SBS | Chr9 | 16781790 | T-A | HET | ||
| SBS | Chr9 | 6893877 | G-A | HET | ||
| SBS | Chr9 | 8358933 | T-G | HET |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 100372 | 100373 | 1 | |
| Deletion | Chr12 | 2800340 | 2800379 | 39 | |
| Deletion | Chr11 | 4683622 | 4683634 | 12 | LOC_Os11g08840 |
| Deletion | Chr5 | 9313022 | 9313025 | 3 | |
| Deletion | Chr9 | 11923800 | 11923803 | 3 | |
| Deletion | Chr3 | 15643911 | 15643915 | 4 | |
| Deletion | Chr10 | 17492362 | 17492368 | 6 | |
| Deletion | Chr8 | 17947733 | 17947739 | 6 | |
| Deletion | Chr10 | 21767436 | 21767437 | 1 | |
| Deletion | Chr11 | 24450313 | 24450319 | 6 | |
| Deletion | Chr4 | 24713489 | 24713490 | 1 | LOC_Os04g41690 |
| Deletion | Chr6 | 27191251 | 27191253 | 2 | LOC_Os06g44960 |
| Deletion | Chr4 | 28661231 | 28661232 | 1 | |
| Deletion | Chr1 | 40795981 | 40795985 | 4 | LOC_Os01g70430 |
| Deletion | Chr1 | 40798280 | 40798292 | 12 | LOC_Os01g70440 |
| Deletion | Chr1 | 40949022 | 40949033 | 11 | LOC_Os01g70740 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 12601954 | 12601956 | 3 | LOC_Os11g21960 |
| Insertion | Chr4 | 26385273 | 26385274 | 2 | |
| Insertion | Chr4 | 26597673 | 26597673 | 1 | |
| Insertion | Chr6 | 5667337 | 5667337 | 1 | |
| Insertion | Chr9 | 12703076 | 12703079 | 4 |
No Inversion
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 1543331 | Chr6 | 20646116 | 2 |
| Translocation | Chr6 | 4360215 | Chr3 | 11111764 | LOC_Os03g19760 |
| Translocation | Chr6 | 4360220 | Chr3 | 11112542 | LOC_Os03g19760 |
| Translocation | Chr5 | 5022236 | Chr2 | 25341897 | LOC_Os02g42134 |
| Translocation | Chr12 | 7742402 | Chr2 | 19414035 | 2 |
| Translocation | Chr6 | 11268343 | Chr3 | 8727276 | 2 |
| Translocation | Chr12 | 12473248 | Chr9 | 5560163 | |
| Translocation | Chr8 | 20427247 | Chr3 | 11117637 | 2 |
| Translocation | Chr8 | 20427258 | Chr3 | 11112549 | 2 |
| Translocation | Chr6 | 22826962 | Chr5 | 15579987 |
