Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3852-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3852-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 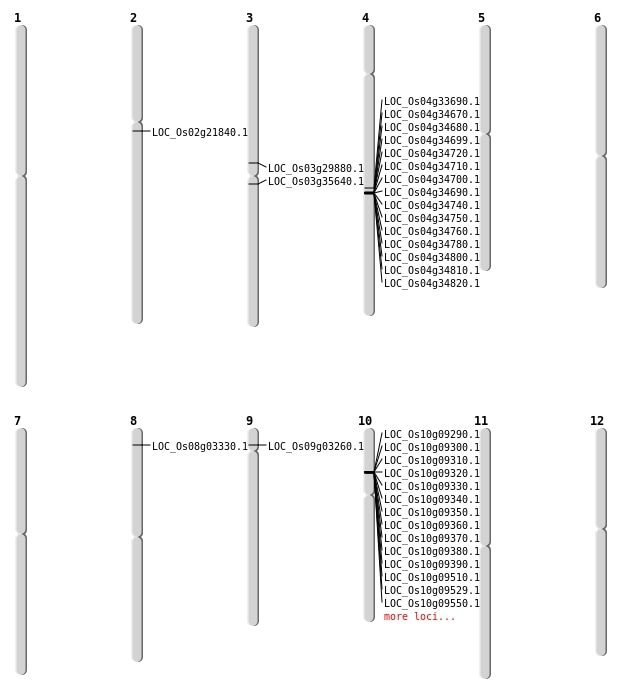
|
| Variant Information |
|---|
Single base substitutions: 48
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12890245 | T-C | Homo | ||
| SBS | Chr1 | 23036150 | G-A | HET | ||
| SBS | Chr1 | 33965735 | T-C | HET | ||
| SBS | Chr1 | 3508465 | G-T | HET | ||
| SBS | Chr10 | 16630819 | C-T | HET | ||
| SBS | Chr10 | 2260722 | C-A | HET | ||
| SBS | Chr10 | 8631515 | G-A | HET | ||
| SBS | Chr11 | 15218675 | C-G | HET | ||
| SBS | Chr11 | 24237317 | T-A | Homo | ||
| SBS | Chr11 | 26982762 | A-G | HET | ||
| SBS | Chr11 | 8615813 | C-T | Homo | ||
| SBS | Chr12 | 12552020 | C-G | HET | ||
| SBS | Chr12 | 3591411 | T-C | HET | ||
| SBS | Chr2 | 1188140 | A-G | Homo | ||
| SBS | Chr2 | 12979946 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g21840 |
| SBS | Chr2 | 13487397 | T-C | HET | ||
| SBS | Chr2 | 20562315 | A-G | HET | ||
| SBS | Chr2 | 34975675 | T-C | HET | ||
| SBS | Chr3 | 11562418 | C-A | HET | ||
| SBS | Chr3 | 14413091 | C-T | Homo | ||
| SBS | Chr3 | 19740705 | G-A | HET | STOP_GAINED | LOC_Os03g35640 |
| SBS | Chr3 | 26651716 | G-A | HET | ||
| SBS | Chr3 | 27438080 | G-A | HET | ||
| SBS | Chr3 | 32192854 | G-A | HET | ||
| SBS | Chr3 | 7129959 | C-T | Homo | ||
| SBS | Chr3 | 9547794 | C-T | Homo | ||
| SBS | Chr4 | 11095608 | G-A | HET | ||
| SBS | Chr4 | 1410033 | T-A | HET | ||
| SBS | Chr4 | 579328 | C-T | HET | ||
| SBS | Chr4 | 98878 | T-C | HET | ||
| SBS | Chr5 | 1032944 | A-C | Homo | ||
| SBS | Chr5 | 10607250 | C-G | Homo | ||
| SBS | Chr5 | 18534623 | C-T | Homo | ||
| SBS | Chr5 | 24448941 | C-A | Homo | ||
| SBS | Chr6 | 12604418 | C-T | HET | ||
| SBS | Chr6 | 14941767 | T-C | HET | ||
| SBS | Chr6 | 29458648 | T-G | HET | ||
| SBS | Chr6 | 901964 | C-T | Homo | ||
| SBS | Chr7 | 11772592 | G-A | HET | ||
| SBS | Chr7 | 15091602 | A-T | HET | ||
| SBS | Chr7 | 15091603 | C-A | HET | ||
| SBS | Chr7 | 4845349 | C-A | HET | ||
| SBS | Chr7 | 8379516 | T-C | HET | ||
| SBS | Chr8 | 11824685 | C-T | HET | ||
| SBS | Chr8 | 1544448 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g03330 |
| SBS | Chr9 | 1568799 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g03260 |
| SBS | Chr9 | 16923834 | G-A | HET | ||
| SBS | Chr9 | 1846002 | G-T | Homo |
Deletions: 12
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 22813662 | 22813662 | 1 | |
| Insertion | Chr2 | 34287786 | 34287789 | 4 | |
| Insertion | Chr3 | 7965487 | 7965488 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 20402349 | 21357565 | LOC_Os04g33690 |
| Inversion | Chr4 | 20991069 | 21107161 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 327627 | Chr4 | 11184837 | |
| Translocation | Chr12 | 5779287 | Chr8 | 19753812 | |
| Translocation | Chr4 | 21357545 | Chr3 | 17028652 | LOC_Os03g29880 |
| Translocation | Chr3 | 29194570 | Chr1 | 31269208 |
