Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3857-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3857-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 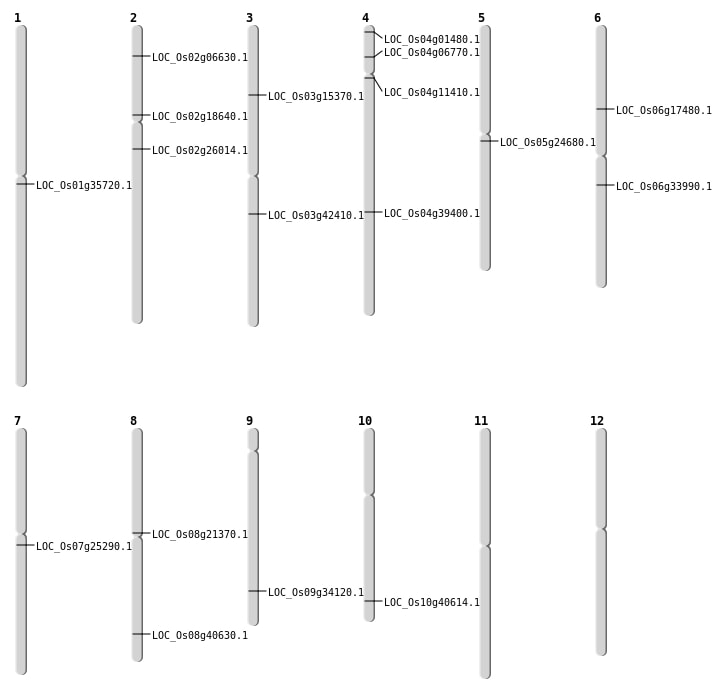
|
| Variant Information |
|---|
Single base substitutions: 48
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11314782 | C-T | HET | ||
| SBS | Chr1 | 18333179 | G-T | HET | ||
| SBS | Chr1 | 19778888 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g35720 |
| SBS | Chr1 | 29848680 | C-T | HET | ||
| SBS | Chr10 | 10160858 | C-T | Homo | ||
| SBS | Chr10 | 12261156 | G-T | Homo | ||
| SBS | Chr10 | 21852885 | A-G | HET | ||
| SBS | Chr12 | 19760487 | T-C | HET | ||
| SBS | Chr12 | 21997777 | T-C | HET | ||
| SBS | Chr12 | 26521361 | G-A | HET | ||
| SBS | Chr12 | 7742721 | C-T | Homo | ||
| SBS | Chr2 | 3336863 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g06630 |
| SBS | Chr2 | 8562485 | T-G | HET | ||
| SBS | Chr3 | 2358939 | T-G | HET | ||
| SBS | Chr3 | 7919417 | G-A | HET | ||
| SBS | Chr4 | 13812445 | C-T | HET | ||
| SBS | Chr4 | 23450822 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g39400 |
| SBS | Chr4 | 328713 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g01480 |
| SBS | Chr4 | 3556207 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g06770 |
| SBS | Chr4 | 7758347 | C-T | HET | ||
| SBS | Chr5 | 10490251 | G-T | HET | ||
| SBS | Chr5 | 10490252 | C-G | HET | ||
| SBS | Chr5 | 14287249 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g24680 |
| SBS | Chr5 | 18386125 | T-G | HET | ||
| SBS | Chr5 | 18422850 | G-C | HET | ||
| SBS | Chr5 | 18594677 | C-A | HET | ||
| SBS | Chr5 | 20572658 | C-A | HET | ||
| SBS | Chr5 | 4473409 | C-G | HET | ||
| SBS | Chr5 | 6443533 | A-G | HET | ||
| SBS | Chr6 | 10138024 | C-A | HET | ||
| SBS | Chr6 | 10138025 | C-T | HET | STOP_GAINED | LOC_Os06g17480 |
| SBS | Chr6 | 13808253 | A-T | HET | ||
| SBS | Chr6 | 13808254 | C-T | HET | ||
| SBS | Chr6 | 19796805 | G-C | HET | STOP_LOST | LOC_Os06g33990 |
| SBS | Chr6 | 22222229 | T-C | HET | ||
| SBS | Chr6 | 2527498 | G-A | Homo | ||
| SBS | Chr6 | 25514594 | G-A | HET | ||
| SBS | Chr7 | 11413747 | C-A | HET | ||
| SBS | Chr7 | 14443266 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g25290 |
| SBS | Chr7 | 15867833 | A-G | Homo | ||
| SBS | Chr7 | 18729214 | T-C | HET | ||
| SBS | Chr8 | 12751493 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g21370 |
| SBS | Chr8 | 17768960 | A-T | HET | ||
| SBS | Chr8 | 20760292 | C-T | HET | ||
| SBS | Chr8 | 22895891 | C-T | HET | ||
| SBS | Chr8 | 27685812 | A-G | Homo | ||
| SBS | Chr9 | 20145943 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g34120 |
| SBS | Chr9 | 5275783 | G-A | Homo |
Deletions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 2038263 | 2038264 | 1 | |
| Deletion | Chr4 | 2797684 | 2797685 | 1 | |
| Deletion | Chr9 | 5031212 | 5031237 | 25 | |
| Deletion | Chr2 | 10862437 | 10862440 | 3 | LOC_Os02g18640 |
| Deletion | Chr4 | 11805866 | 11805867 | 1 | |
| Deletion | Chr4 | 12527674 | 12527675 | 1 | |
| Deletion | Chr9 | 16675267 | 16675274 | 7 | |
| Deletion | Chr4 | 17398568 | 17398577 | 9 | |
| Deletion | Chr11 | 19627359 | 19627360 | 1 | |
| Deletion | Chr5 | 21095691 | 21095697 | 6 | |
| Deletion | Chr10 | 21786478 | 21786491 | 13 | LOC_Os10g40614 |
| Deletion | Chr9 | 22148296 | 22148299 | 3 | |
| Deletion | Chr3 | 23597198 | 23597199 | 1 | LOC_Os03g42410 |
| Deletion | Chr8 | 25721096 | 25721101 | 5 | LOC_Os08g40630 |
| Deletion | Chr2 | 26418737 | 26418749 | 12 |
Insertions: 5
No Inversion
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 7917874 | Chr3 | 8412207 | LOC_Os03g15370 |
| Translocation | Chr5 | 9090903 | Chr2 | 15277489 | LOC_Os02g26014 |
| Translocation | Chr12 | 13545259 | Chr4 | 6233521 | LOC_Os04g11410 |
| Translocation | Chr7 | 20170939 | Chr6 | 5157867 |
