Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3858-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3858-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 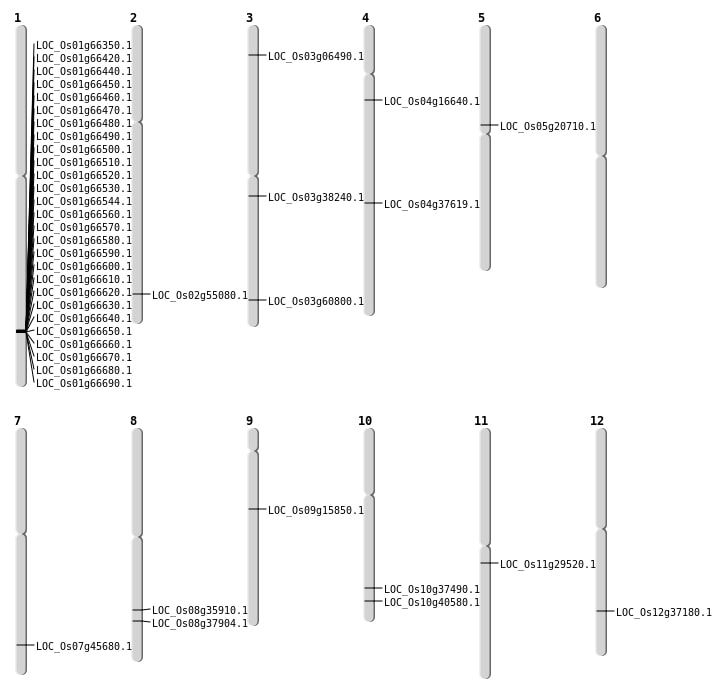
|
| Variant Information |
|---|
Single base substitutions: 59
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18184207 | A-T | HET | ||
| SBS | Chr1 | 19806448 | G-T | HET | ||
| SBS | Chr1 | 2284226 | G-A | Homo | ||
| SBS | Chr1 | 34456046 | G-C | HET | ||
| SBS | Chr1 | 34784068 | C-T | Homo | ||
| SBS | Chr1 | 36848216 | T-C | Homo | ||
| SBS | Chr1 | 464454 | G-A | HET | ||
| SBS | Chr1 | 5585933 | G-A | HET | ||
| SBS | Chr10 | 13776831 | T-C | HET | ||
| SBS | Chr10 | 19822752 | C-A | HET | ||
| SBS | Chr10 | 20073128 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g37490 |
| SBS | Chr10 | 4608837 | T-C | Homo | ||
| SBS | Chr11 | 10292514 | C-A | HET | ||
| SBS | Chr11 | 17129610 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g29520 |
| SBS | Chr11 | 1810289 | G-A | HET | ||
| SBS | Chr11 | 18548986 | T-C | HET | ||
| SBS | Chr11 | 26088359 | G-T | HET | ||
| SBS | Chr11 | 2858468 | G-A | HET | ||
| SBS | Chr12 | 9872254 | G-A | Homo | ||
| SBS | Chr2 | 11717785 | G-A | HET | ||
| SBS | Chr2 | 19061654 | G-T | HET | ||
| SBS | Chr2 | 4632130 | G-T | HET | ||
| SBS | Chr2 | 7338455 | C-T | HET | ||
| SBS | Chr3 | 21229437 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g38240 |
| SBS | Chr3 | 28116517 | C-T | HET | ||
| SBS | Chr3 | 28169116 | C-T | HET | ||
| SBS | Chr3 | 34544864 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g60800 |
| SBS | Chr4 | 11147939 | C-T | HET | ||
| SBS | Chr4 | 21130693 | G-A | HET | ||
| SBS | Chr4 | 21534660 | C-T | HET | ||
| SBS | Chr4 | 30831994 | A-T | HET | ||
| SBS | Chr4 | 35325971 | C-A | HET | ||
| SBS | Chr4 | 3961686 | A-T | Homo | ||
| SBS | Chr4 | 4375829 | A-T | Homo | ||
| SBS | Chr4 | 8545147 | C-T | HET | ||
| SBS | Chr4 | 9052267 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g16640 |
| SBS | Chr5 | 1047964 | A-G | HET | ||
| SBS | Chr5 | 12140692 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g20710 |
| SBS | Chr5 | 13595853 | C-G | HET | ||
| SBS | Chr5 | 16356302 | C-G | HET | ||
| SBS | Chr5 | 1724178 | G-T | HET | ||
| SBS | Chr5 | 27386558 | A-T | HET | ||
| SBS | Chr5 | 29402946 | A-T | Homo | ||
| SBS | Chr5 | 29454636 | C-T | Homo | ||
| SBS | Chr6 | 19918405 | T-C | HET | ||
| SBS | Chr6 | 5518211 | G-A | HET | ||
| SBS | Chr6 | 7650055 | C-T | HET | ||
| SBS | Chr7 | 10578403 | C-T | HET | ||
| SBS | Chr7 | 12409631 | G-A | HET | ||
| SBS | Chr7 | 17704890 | G-A | HET | ||
| SBS | Chr7 | 19420700 | T-A | HET | ||
| SBS | Chr7 | 27275221 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g45680 |
| SBS | Chr7 | 9542651 | A-G | HET | ||
| SBS | Chr7 | 9834261 | G-A | HET | ||
| SBS | Chr8 | 24007976 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g37904 |
| SBS | Chr9 | 13664657 | T-C | HET | ||
| SBS | Chr9 | 14874622 | G-A | HET | ||
| SBS | Chr9 | 15903374 | C-T | HET | ||
| SBS | Chr9 | 3762542 | T-G | HET |
Deletions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 1815203 | 1815206 | 3 | |
| Deletion | Chr3 | 2957119 | 2957141 | 22 | |
| Deletion | Chr4 | 4406576 | 4406579 | 3 | |
| Deletion | Chr11 | 6449375 | 6449376 | 1 | |
| Deletion | Chr9 | 13387559 | 13387562 | 3 | |
| Deletion | Chr12 | 25333799 | 25333816 | 17 | |
| Deletion | Chr5 | 29161923 | 29161925 | 2 | |
| Deletion | Chr2 | 33735469 | 33735470 | 1 | LOC_Os02g55080 |
| Deletion | Chr3 | 34104202 | 34104203 | 1 | |
| Deletion | Chr4 | 34504049 | 34504053 | 4 | |
| Deletion | Chr1 | 38563001 | 38739000 | 175999 | 26 |
Insertions: 6
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 8617829 | 9217240 | |
| Inversion | Chr9 | 9156296 | 9687166 | LOC_Os09g15850 |
| Inversion | Chr9 | 9156307 | 9687166 | LOC_Os09g15850 |
| Inversion | Chr4 | 22370573 | 22708253 | LOC_Os04g37619 |
| Inversion | Chr1 | 38404832 | 38534961 | LOC_Os01g66350 |
| Inversion | Chr1 | 38404846 | 38534970 | LOC_Os01g66350 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 5979547 | Chr6 | 10020348 | |
| Translocation | Chr6 | 7792043 | Chr3 | 3255331 | LOC_Os03g06490 |
| Translocation | Chr6 | 7793186 | Chr3 | 3255320 | LOC_Os03g06490 |
| Translocation | Chr9 | 11971554 | Chr2 | 24766488 | |
| Translocation | Chr10 | 13367222 | Chr1 | 25286864 | |
| Translocation | Chr10 | 21733985 | Chr2 | 18287300 | LOC_Os10g40580 |
| Translocation | Chr8 | 22651598 | Chr7 | 3872019 | LOC_Os08g35910 |
| Translocation | Chr12 | 22800140 | Chr9 | 4757324 | LOC_Os12g37180 |
