Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3885-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3885-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 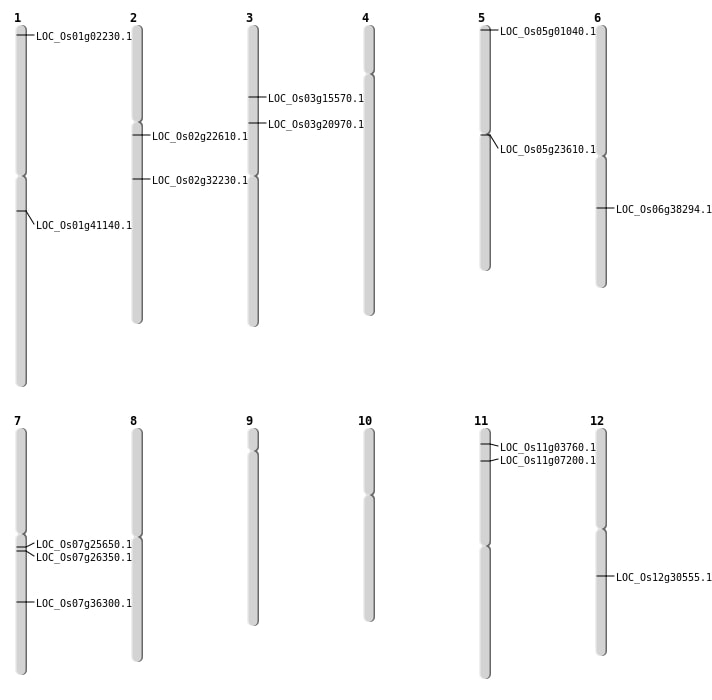
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12640088 | G-A | Homo | ||
| SBS | Chr1 | 13432934 | A-T | Homo | ||
| SBS | Chr1 | 15377433 | C-A | Homo | ||
| SBS | Chr1 | 23292218 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g41140 |
| SBS | Chr1 | 34265895 | G-A | HET | ||
| SBS | Chr1 | 36203491 | C-T | HET | ||
| SBS | Chr1 | 36417518 | G-A | HET | ||
| SBS | Chr10 | 3028995 | T-C | Homo | ||
| SBS | Chr12 | 6787358 | C-T | HET | ||
| SBS | Chr12 | 885166 | C-T | HET | ||
| SBS | Chr2 | 19039074 | A-T | HET | STOP_GAINED | LOC_Os02g32230 |
| SBS | Chr2 | 8840587 | A-G | Homo | ||
| SBS | Chr3 | 23242290 | T-C | HET | ||
| SBS | Chr3 | 24666764 | G-A | HET | ||
| SBS | Chr3 | 26188478 | A-C | HET | ||
| SBS | Chr3 | 8573919 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g15570 |
| SBS | Chr5 | 22103171 | G-T | HET | ||
| SBS | Chr5 | 22829214 | A-G | Homo | ||
| SBS | Chr5 | 29614589 | C-T | HET | ||
| SBS | Chr5 | 69525 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g01040 |
| SBS | Chr6 | 1147755 | A-G | Homo | ||
| SBS | Chr6 | 27762771 | G-T | HET | ||
| SBS | Chr7 | 5140290 | T-A | Homo | ||
| SBS | Chr7 | 5140291 | T-A | Homo | ||
| SBS | Chr8 | 17163550 | C-T | Homo | ||
| SBS | Chr9 | 16986618 | A-G | Homo | ||
| SBS | Chr9 | 9842720 | T-C | Homo |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 247569 | 247583 | 14 | |
| Deletion | Chr11 | 423315 | 423330 | 15 | |
| Deletion | Chr1 | 668156 | 668160 | 4 | LOC_Os01g02230 |
| Deletion | Chr6 | 1191284 | 1191297 | 13 | |
| Deletion | Chr4 | 1219970 | 1219973 | 3 | |
| Deletion | Chr10 | 2078301 | 2078302 | 1 | |
| Deletion | Chr9 | 4508778 | 4508780 | 2 | |
| Deletion | Chr2 | 6422058 | 6422061 | 3 | |
| Deletion | Chr3 | 6987739 | 6987740 | 1 | |
| Deletion | Chr7 | 9992583 | 9992588 | 5 | |
| Deletion | Chr1 | 10454974 | 10454985 | 11 | |
| Deletion | Chr4 | 10977870 | 10977873 | 3 | |
| Deletion | Chr3 | 11894728 | 11894729 | 1 | LOC_Os03g20970 |
| Deletion | Chr5 | 11896510 | 11896512 | 2 | |
| Deletion | Chr4 | 11939375 | 11939388 | 13 | |
| Deletion | Chr5 | 12131073 | 12131074 | 1 | |
| Deletion | Chr4 | 12781542 | 12781545 | 3 | |
| Deletion | Chr2 | 15334931 | 15334932 | 1 | |
| Deletion | Chr4 | 25514677 | 25514692 | 15 | |
| Deletion | Chr11 | 28699148 | 28699149 | 1 | |
| Deletion | Chr2 | 33970881 | 33970885 | 4 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 10480745 | 10480745 | 1 | |
| Insertion | Chr11 | 22549521 | 22549746 | 226 | |
| Insertion | Chr3 | 17537746 | 17537803 | 58 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 5768893 | 5769163 | |
| Inversion | Chr5 | 12872860 | 13601677 | |
| Inversion | Chr7 | 14703455 | 15133295 | LOC_Os07g25650 |
| Inversion | Chr7 | 15133303 | 15145552 | LOC_Os07g26350 |
| Inversion | Chr8 | 21898140 | 21898448 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 3612989 | Chr7 | 26946719 | LOC_Os11g07200 |
| Translocation | Chr12 | 7608871 | Chr2 | 13477787 | LOC_Os02g22610 |
| Translocation | Chr10 | 7674106 | Chr3 | 25685778 | |
| Translocation | Chr9 | 12085432 | Chr6 | 25881774 | |
| Translocation | Chr12 | 18355317 | Chr11 | 1476168 | 2 |
| Translocation | Chr11 | 19390373 | Chr2 | 23245125 | |
| Translocation | Chr7 | 21711356 | Chr5 | 13540457 | 2 |
| Translocation | Chr6 | 22658470 | Chr1 | 299895 | LOC_Os06g38294 |
| Translocation | Chr11 | 24374256 | Chr8 | 17449731 |
