Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN391-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN391-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 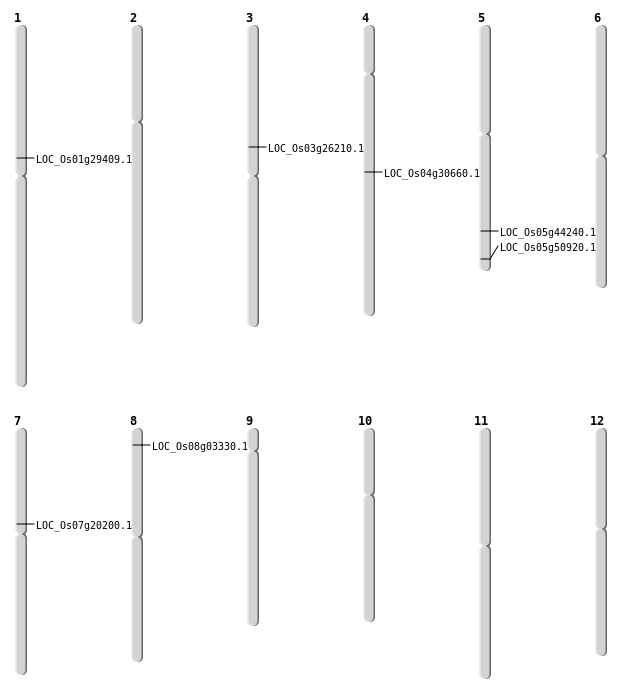
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12324754 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 29594985 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 43074558 | A-T | HET | INTRON | |
| SBS | Chr1 | 8954081 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 8954086 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 13996322 | T-G | HET | INTERGENIC | |
| SBS | Chr10 | 16104486 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 5727285 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 8911839 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 12683377 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 24237317 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 6289819 | A-C | HOMO | INTRON | |
| SBS | Chr12 | 14794985 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 2798497 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 3006927 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 18544458 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 24252908 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 14413091 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 15000890 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g26210 |
| SBS | Chr3 | 20256583 | G-A | HET | INTRON | |
| SBS | Chr3 | 7129959 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 13074275 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 13720879 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 24448941 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 25734424 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g44240 |
| SBS | Chr5 | 4862221 | A-G | HET | INTRON | |
| SBS | Chr5 | 5379369 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 24869242 | T-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 11658994 | G-T | HOMO | STOP_GAINED | LOC_Os07g20200 |
| SBS | Chr8 | 13091428 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 1544448 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g03330 |
| SBS | Chr8 | 3888824 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 4295690 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 5827168 | G-A | HET | UTR_3_PRIME |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 1711358 | 1711368 | 10 | |
| Deletion | Chr7 | 2804425 | 2804430 | 5 | |
| Deletion | Chr5 | 3466387 | 3466392 | 5 | |
| Deletion | Chr4 | 4051112 | 4051114 | 2 | |
| Deletion | Chr11 | 7849673 | 7849683 | 10 | |
| Deletion | Chr7 | 8984896 | 8984897 | 1 | |
| Deletion | Chr7 | 9193067 | 9193090 | 23 | |
| Deletion | Chr11 | 9412195 | 9412196 | 1 | |
| Deletion | Chr9 | 9588698 | 9588699 | 1 | |
| Deletion | Chr10 | 10568634 | 10568638 | 4 | |
| Deletion | Chr1 | 11931981 | 11932021 | 40 | |
| Deletion | Chr11 | 12680360 | 12680373 | 13 | |
| Deletion | Chr5 | 14206712 | 14206720 | 8 | |
| Deletion | Chr1 | 16493524 | 16493528 | 4 | LOC_Os01g29409 |
| Deletion | Chr8 | 16922091 | 16922092 | 1 | |
| Deletion | Chr4 | 18319973 | 18319976 | 3 | LOC_Os04g30660 |
| Deletion | Chr4 | 18808711 | 18808712 | 1 | |
| Deletion | Chr11 | 20487234 | 20487236 | 2 | |
| Deletion | Chr11 | 20622834 | 20622844 | 10 | |
| Deletion | Chr10 | 21654437 | 21654441 | 4 | |
| Deletion | Chr8 | 22135462 | 22135478 | 16 | |
| Deletion | Chr10 | 22557369 | 22557371 | 2 | |
| Deletion | Chr5 | 24503154 | 24503159 | 5 | |
| Deletion | Chr5 | 24518239 | 24518241 | 2 | |
| Deletion | Chr5 | 25508645 | 25508656 | 11 | |
| Deletion | Chr6 | 25720753 | 25720763 | 10 | |
| Deletion | Chr11 | 25763803 | 25763804 | 1 | |
| Deletion | Chr6 | 28548078 | 28548093 | 15 | |
| Deletion | Chr5 | 29223650 | 29223651 | 1 | LOC_Os05g50920 |
| Deletion | Chr2 | 29535381 | 29535391 | 10 | |
| Deletion | Chr1 | 37394001 | 37394002 | 1 |
Insertions: 8
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 11616744 | 29505948 | |
| Inversion | Chr5 | 27738935 | 29505940 |
No Translocation
