Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3913-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3913-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 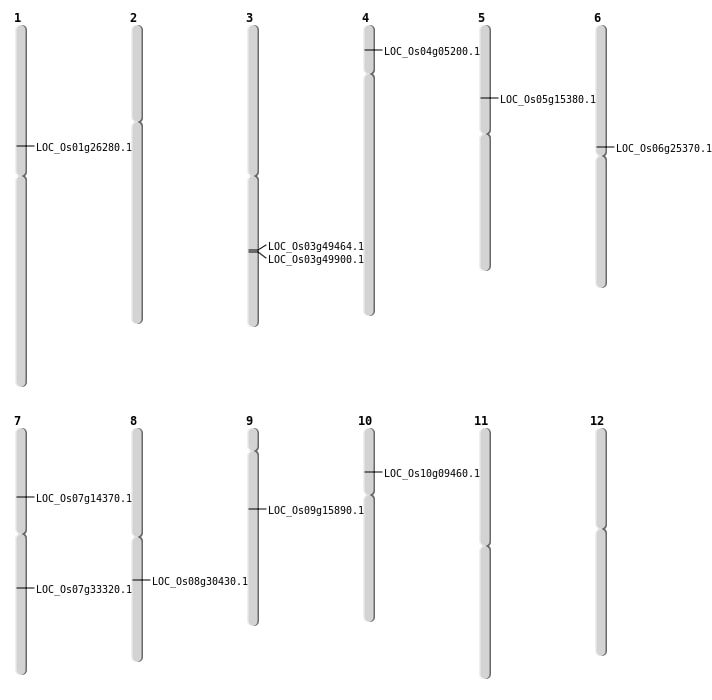
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14539704 | C-G | HET | ||
| SBS | Chr1 | 14898466 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g26280 |
| SBS | Chr1 | 17766329 | A-T | HET | ||
| SBS | Chr1 | 21045141 | A-T | HET | ||
| SBS | Chr10 | 14333903 | C-A | HET | ||
| SBS | Chr10 | 3398093 | G-T | HET | ||
| SBS | Chr10 | 3961054 | G-A | HET | ||
| SBS | Chr11 | 10783010 | T-G | HET | ||
| SBS | Chr12 | 20056054 | A-G | Homo | ||
| SBS | Chr12 | 22028468 | T-A | HET | ||
| SBS | Chr3 | 1180640 | C-A | HET | ||
| SBS | Chr3 | 31984855 | C-T | HET | ||
| SBS | Chr4 | 13532708 | C-T | HET | ||
| SBS | Chr4 | 2594283 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g05200 |
| SBS | Chr4 | 6102002 | C-A | HET | ||
| SBS | Chr5 | 8706149 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g15380 |
| SBS | Chr6 | 14860784 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g25370 |
| SBS | Chr6 | 29011517 | A-T | HET | ||
| SBS | Chr6 | 30324369 | T-C | HET | ||
| SBS | Chr7 | 10865679 | C-T | HET | ||
| SBS | Chr7 | 18727299 | G-C | HET | ||
| SBS | Chr7 | 2425728 | G-T | HET | ||
| SBS | Chr8 | 15800427 | A-G | HET | ||
| SBS | Chr8 | 17002407 | G-A | HET | ||
| SBS | Chr8 | 17532393 | G-A | HET | ||
| SBS | Chr8 | 18785311 | A-G | HET | ||
| SBS | Chr8 | 9728489 | C-T | HET | ||
| SBS | Chr9 | 6525726 | C-T | HET | ||
| SBS | Chr9 | 7491172 | C-T | HET | ||
| SBS | Chr9 | 9470411 | A-G | HET | ||
| SBS | Chr9 | 9710730 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g15890 |
Deletions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 5101914 | 5101915 | 1 | LOC_Os10g09460 |
| Deletion | Chr7 | 8206813 | 8206816 | 3 | LOC_Os07g14370 |
| Deletion | Chr1 | 9958174 | 9958175 | 1 | |
| Deletion | Chr5 | 11918696 | 11918699 | 3 | |
| Deletion | Chr10 | 12893630 | 12893637 | 7 | |
| Deletion | Chr11 | 14829012 | 14829014 | 2 | |
| Deletion | Chr10 | 15105749 | 15105750 | 1 | |
| Deletion | Chr4 | 15283580 | 15283581 | 1 | |
| Deletion | Chr2 | 15534797 | 15534798 | 1 | |
| Deletion | Chr8 | 18730411 | 18730439 | 28 | LOC_Os08g30430 |
| Deletion | Chr2 | 22305112 | 22305115 | 3 | |
| Deletion | Chr12 | 23534993 | 23534997 | 4 | |
| Deletion | Chr7 | 25051177 | 25051179 | 2 | |
| Deletion | Chr3 | 32333855 | 32333871 | 16 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 14849652 | 14849653 | 2 | |
| Insertion | Chr3 | 5652319 | 5652319 | 1 | |
| Insertion | Chr7 | 21095996 | 21095997 | 2 | |
| Insertion | Chr8 | 9188935 | 9188936 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 19509482 | 19916986 | LOC_Os07g33320 |
| Inversion | Chr7 | 19509484 | 19917000 | LOC_Os07g33320 |
| Inversion | Chr3 | 28156568 | 28453533 | 2 |
| Inversion | Chr3 | 28156572 | 28453546 | 2 |
No Translocation
