Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3916-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3916-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 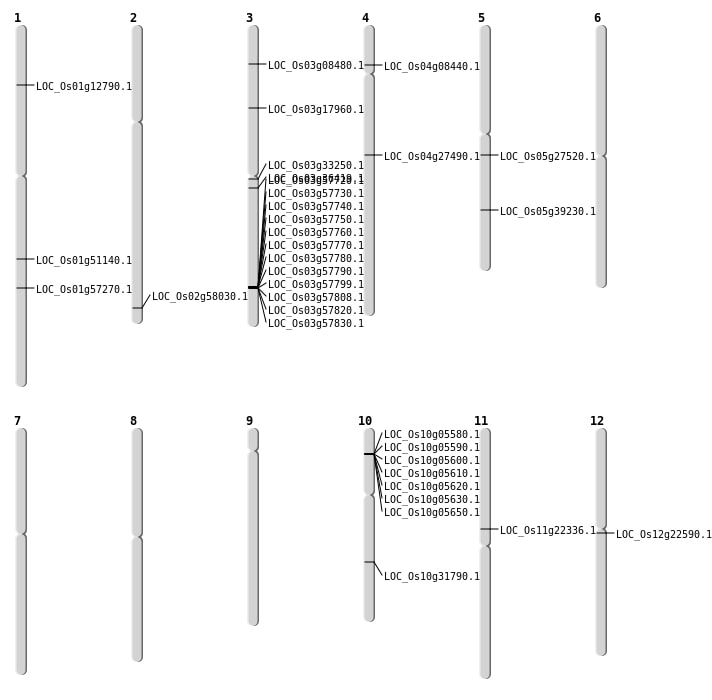
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11855145 | A-G | HET | ||
| SBS | Chr1 | 14333821 | C-T | HET | ||
| SBS | Chr1 | 7084378 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g12790 |
| SBS | Chr10 | 21820242 | C-T | HET | ||
| SBS | Chr10 | 37619 | G-A | HET | ||
| SBS | Chr11 | 12803556 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g22336 |
| SBS | Chr11 | 14780400 | G-A | HET | ||
| SBS | Chr11 | 6389658 | A-C | HET | ||
| SBS | Chr12 | 21628505 | C-T | HET | ||
| SBS | Chr12 | 23544543 | C-A | HET | ||
| SBS | Chr12 | 25618887 | C-A | HET | ||
| SBS | Chr2 | 31241509 | C-T | HET | ||
| SBS | Chr3 | 10001685 | C-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g17960 |
| SBS | Chr3 | 1242000 | G-A | Homo | ||
| SBS | Chr3 | 20187127 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g36419 |
| SBS | Chr3 | 20187128 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g36419 |
| SBS | Chr3 | 20779423 | G-A | Homo | ||
| SBS | Chr3 | 28355359 | G-T | Homo | ||
| SBS | Chr3 | 4360133 | T-A | HET | STOP_GAINED | LOC_Os03g08480 |
| SBS | Chr3 | 6006875 | C-T | HET | ||
| SBS | Chr4 | 16250695 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g27490 |
| SBS | Chr4 | 34266962 | G-A | HET | ||
| SBS | Chr4 | 4533797 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g08440 |
| SBS | Chr4 | 9081809 | G-C | HET | ||
| SBS | Chr4 | 9631447 | C-T | HET | ||
| SBS | Chr5 | 16009713 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g27520 |
| SBS | Chr5 | 17332862 | A-G | HET | ||
| SBS | Chr5 | 17579643 | C-T | HET | ||
| SBS | Chr5 | 27428050 | G-T | HET | ||
| SBS | Chr6 | 12214809 | G-A | HET | ||
| SBS | Chr6 | 27916476 | T-C | HET | ||
| SBS | Chr6 | 6734297 | C-T | HET | ||
| SBS | Chr7 | 13188547 | G-A | Homo | ||
| SBS | Chr8 | 14079423 | C-A | Homo | ||
| SBS | Chr8 | 16639662 | A-G | HET | ||
| SBS | Chr8 | 25704721 | G-T | HET | ||
| SBS | Chr9 | 4332497 | G-T | HET | ||
| SBS | Chr9 | 6197893 | G-T | HET | ||
| SBS | Chr9 | 893647 | G-A | HET |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 1509201 | 1509207 | 6 | |
| Deletion | Chr10 | 2774001 | 2822000 | 47999 | 7 |
| Deletion | Chr3 | 4128219 | 4128224 | 5 | |
| Deletion | Chr4 | 6599611 | 6599612 | 1 | |
| Deletion | Chr9 | 8963674 | 8963688 | 14 | |
| Deletion | Chr12 | 12748236 | 12748262 | 26 | LOC_Os12g22590 |
| Deletion | Chr8 | 15171857 | 15171948 | 91 | |
| Deletion | Chr10 | 16320446 | 16320460 | 14 | |
| Deletion | Chr5 | 16535092 | 16535101 | 9 | |
| Deletion | Chr8 | 17058106 | 17058156 | 50 | |
| Deletion | Chr3 | 19023370 | 19023374 | 4 | LOC_Os03g33250 |
| Deletion | Chr9 | 19804797 | 19804799 | 2 | |
| Deletion | Chr5 | 23004744 | 23004751 | 7 | LOC_Os05g39230 |
| Deletion | Chr4 | 26264165 | 26264177 | 12 | |
| Deletion | Chr12 | 26403639 | 26403646 | 7 | |
| Deletion | Chr2 | 29236697 | 29236702 | 5 | |
| Deletion | Chr3 | 32888001 | 32951000 | 62999 | 12 |
| Deletion | Chr1 | 33095299 | 33095329 | 30 | LOC_Os01g57270 |
Insertions: 8
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 9265589 | 9834435 | |
| Inversion | Chr11 | 9265632 | 9834447 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 16667203 | Chr2 | 35527139 | 2 |
| Translocation | Chr10 | 16667206 | Chr2 | 35526853 | 2 |
| Translocation | Chr3 | 22761146 | Chr1 | 29385505 | LOC_Os01g51140 |
| Translocation | Chr6 | 23392641 | Chr2 | 20261349 | |
| Translocation | Chr6 | 23454674 | Chr2 | 19681247 |
