Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3937-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3937-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 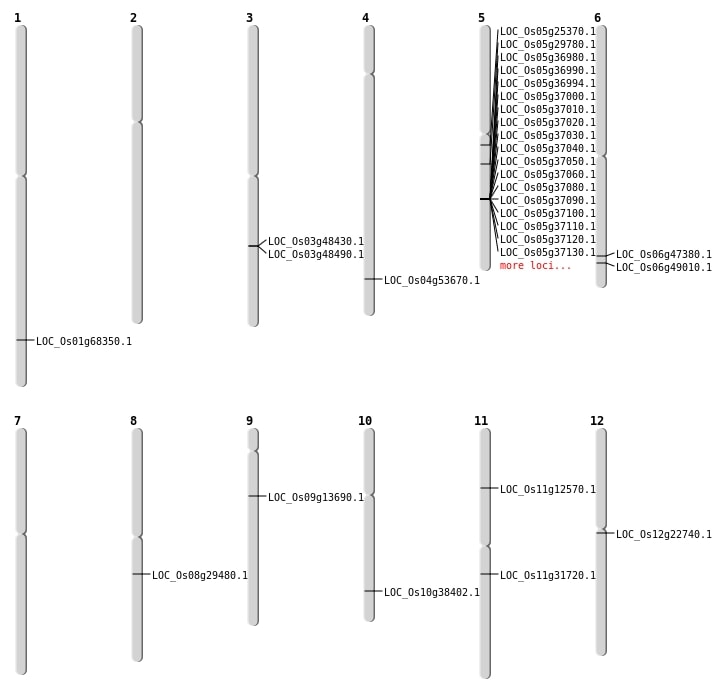
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15212172 | A-G | HET | ||
| SBS | Chr1 | 23149454 | G-C | HET | ||
| SBS | Chr1 | 3568384 | T-G | Homo | ||
| SBS | Chr1 | 35763383 | A-T | Homo | ||
| SBS | Chr1 | 35763384 | A-T | Homo | ||
| SBS | Chr1 | 39716535 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g68350 |
| SBS | Chr10 | 1895170 | G-A | Homo | ||
| SBS | Chr11 | 10308149 | C-T | HET | ||
| SBS | Chr11 | 18601882 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g31720 |
| SBS | Chr11 | 23763763 | G-A | Homo | ||
| SBS | Chr11 | 7066250 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g12570 |
| SBS | Chr11 | 8235549 | C-T | HET | ||
| SBS | Chr12 | 11174556 | G-A | Homo | ||
| SBS | Chr12 | 1431259 | A-G | HET | ||
| SBS | Chr12 | 27467791 | G-A | HET | ||
| SBS | Chr2 | 20078481 | C-T | HET | ||
| SBS | Chr2 | 35100894 | A-T | Homo | ||
| SBS | Chr3 | 27039948 | G-A | HET | ||
| SBS | Chr5 | 2393919 | G-A | HET | ||
| SBS | Chr5 | 27541819 | A-G | HET | ||
| SBS | Chr5 | 4794471 | C-T | HET | ||
| SBS | Chr5 | 5674208 | A-C | HET | ||
| SBS | Chr5 | 8194430 | A-G | HET | ||
| SBS | Chr6 | 19919977 | C-T | HET | ||
| SBS | Chr6 | 24180715 | C-T | Homo | ||
| SBS | Chr6 | 29705458 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g49010 |
| SBS | Chr7 | 25307995 | G-T | HET | ||
| SBS | Chr7 | 25889590 | G-T | HET | ||
| SBS | Chr8 | 18012873 | T-A | Homo | ||
| SBS | Chr8 | 7516616 | T-C | Homo | ||
| SBS | Chr9 | 8006784 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g13690 |
| SBS | Chr9 | 8906642 | G-A | HET |
Deletions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 2535995 | 2535996 | 1 | |
| Deletion | Chr5 | 21603001 | 21658000 | 54999 | 10 |
| Deletion | Chr5 | 21666001 | 22150000 | 483999 | 72 |
| Deletion | Chr12 | 23273082 | 23273083 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr3 | 20923386 | 20923387 | 2 | |
| Insertion | Chr4 | 31847295 | 31847295 | 1 | |
| Insertion | Chr6 | 28719152 | 28719157 | 6 | LOC_Os06g47380 |
| Insertion | Chr7 | 8338880 | 8338881 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 27580959 | 27652240 | 2 |
| Inversion | Chr3 | 27580983 | 27652246 | 2 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 5537862 | Chr4 | 8208165 | |
| Translocation | Chr12 | 12830408 | Chr10 | 7615244 | LOC_Os12g22740 |
| Translocation | Chr12 | 12830923 | Chr10 | 7615240 | LOC_Os12g22740 |
| Translocation | Chr11 | 13306791 | Chr4 | 31987972 | LOC_Os04g53670 |
| Translocation | Chr8 | 13370126 | Chr4 | 2808013 | |
| Translocation | Chr11 | 15594337 | Chr3 | 21568112 | |
| Translocation | Chr8 | 18071283 | Chr5 | 17231038 | 2 |
| Translocation | Chr10 | 20517832 | Chr7 | 11825755 | LOC_Os10g38402 |
| Translocation | Chr12 | 24881283 | Chr5 | 14734872 | LOC_Os05g25370 |
