Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN404-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN404-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 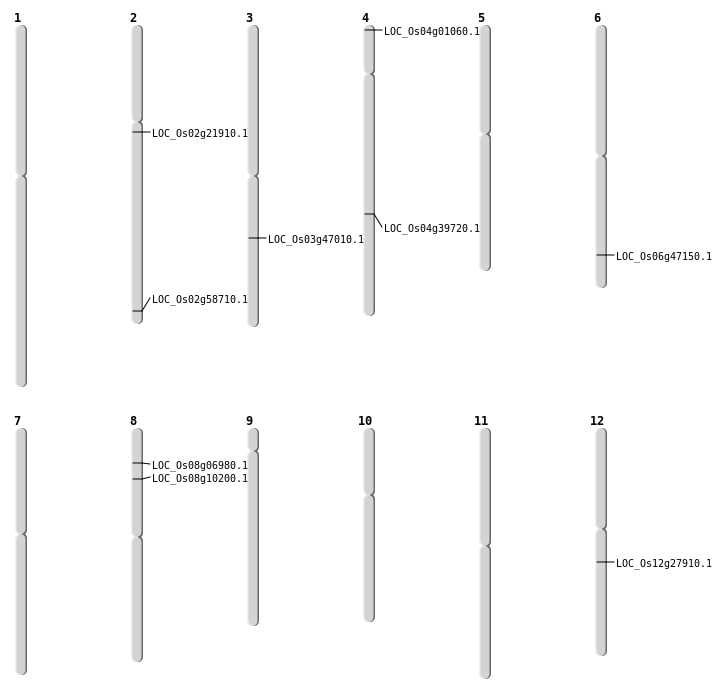
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12364883 | G-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 20177577 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 22597169 | G-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 2844904 | C-G | HET | INTRON | |
| SBS | Chr1 | 3733798 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 15429260 | T-C | HET | INTRON | |
| SBS | Chr10 | 3388220 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 19058277 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 3487904 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 16456429 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g27910 |
| SBS | Chr12 | 16456430 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g27910 |
| SBS | Chr12 | 21162305 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 13027569 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g21910 |
| SBS | Chr2 | 30621811 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 31328247 | C-T | HET | SYNONYMOUS_STOP | |
| SBS | Chr2 | 35869787 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g58710 |
| SBS | Chr3 | 18112659 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 26187772 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 15110802 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 25855172 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 2806330 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 295662 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 31708417 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 3829756 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 52352 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g01060 |
| SBS | Chr4 | 5687399 | A-G | HET | INTRON | |
| SBS | Chr4 | 602723 | A-C | HET | INTRON | |
| SBS | Chr5 | 12942152 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 28701834 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 29081311 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 15572578 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 11084430 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 13259781 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 12485514 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 12485515 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 3887433 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g06980 |
| SBS | Chr8 | 5913693 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g10200 |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 972657 | 972672 | 15 | |
| Deletion | Chr10 | 3009829 | 3009835 | 6 | |
| Deletion | Chr9 | 3743978 | 3743991 | 13 | |
| Deletion | Chr1 | 3814793 | 3814815 | 22 | |
| Deletion | Chr6 | 13337189 | 13337198 | 9 | |
| Deletion | Chr6 | 13409686 | 13409687 | 1 | |
| Deletion | Chr10 | 13591883 | 13591900 | 17 | |
| Deletion | Chr5 | 13759197 | 13759198 | 1 | |
| Deletion | Chr3 | 13769810 | 13769813 | 3 | |
| Deletion | Chr10 | 13860609 | 13860615 | 6 | |
| Deletion | Chr12 | 16904766 | 16904768 | 2 | |
| Deletion | Chr6 | 20641607 | 20641608 | 1 | |
| Deletion | Chr4 | 23672349 | 23672364 | 15 | LOC_Os04g39720 |
| Deletion | Chr7 | 23832840 | 23832841 | 1 | |
| Deletion | Chr7 | 24907243 | 24907255 | 12 | |
| Deletion | Chr2 | 26165278 | 26165279 | 1 | |
| Deletion | Chr3 | 26577512 | 26577528 | 16 | LOC_Os03g47010 |
| Deletion | Chr6 | 28588207 | 28588216 | 9 | LOC_Os06g47150 |
| Deletion | Chr1 | 40731147 | 40731156 | 9 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 4807434 | 4807435 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 30359938 | 30455653 | |
| Inversion | Chr4 | 31711142 | 31759736 |
No Translocation
