Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4111-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4111-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 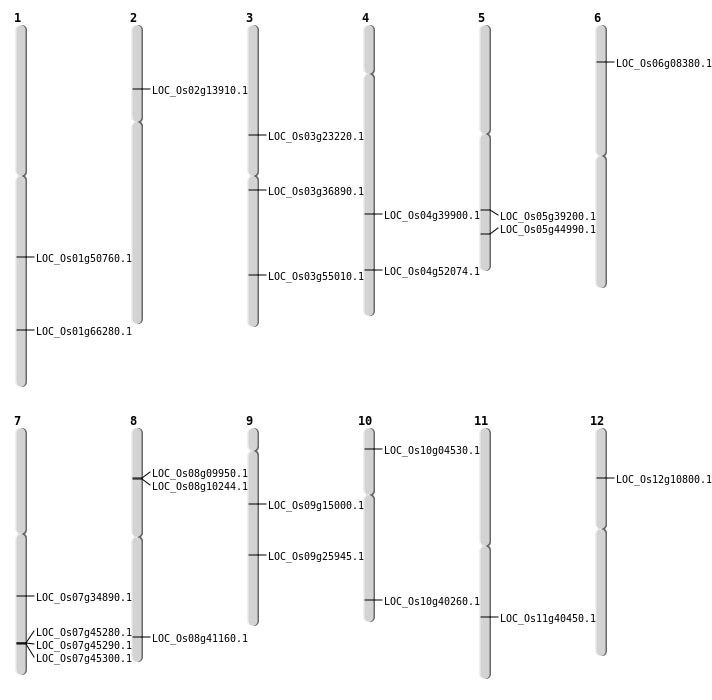
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 29160244 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g50760 |
| SBS | Chr1 | 29160245 | A-T | Homo | ||
| SBS | Chr1 | 33036573 | G-A | HET | ||
| SBS | Chr1 | 43048045 | G-A | HET | ||
| SBS | Chr10 | 13901945 | C-A | HET | ||
| SBS | Chr10 | 16760269 | G-A | Homo | ||
| SBS | Chr10 | 2149198 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g04530 |
| SBS | Chr11 | 19034996 | A-T | HET | ||
| SBS | Chr11 | 5391184 | T-A | HET | ||
| SBS | Chr12 | 21804016 | G-A | HET | ||
| SBS | Chr12 | 24713650 | C-A | HET | ||
| SBS | Chr12 | 368680 | C-G | HET | ||
| SBS | Chr12 | 5808305 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g10800 |
| SBS | Chr3 | 13439272 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g23220 |
| SBS | Chr3 | 20467536 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g36890 |
| SBS | Chr3 | 30286682 | C-T | Homo | ||
| SBS | Chr4 | 33741820 | G-A | HET | ||
| SBS | Chr4 | 35346294 | C-T | Homo | ||
| SBS | Chr4 | 35460721 | T-C | Homo | ||
| SBS | Chr4 | 643790 | A-T | HET | ||
| SBS | Chr5 | 5062678 | T-G | HET | ||
| SBS | Chr6 | 27074693 | G-T | HET | ||
| SBS | Chr7 | 11000119 | C-A | HET | ||
| SBS | Chr7 | 16904325 | C-T | Homo | ||
| SBS | Chr7 | 20912311 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g34890 |
| SBS | Chr7 | 22599846 | G-A | HET | ||
| SBS | Chr7 | 27501365 | T-C | HET | ||
| SBS | Chr8 | 12024379 | G-C | HET | ||
| SBS | Chr8 | 21750557 | C-T | Homo | ||
| SBS | Chr8 | 2245564 | A-G | HET | ||
| SBS | Chr9 | 10203026 | G-A | HET | ||
| SBS | Chr9 | 14941671 | G-A | HET | ||
| SBS | Chr9 | 15582297 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g25945 |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 2119390 | 2119391 | 1 | |
| Deletion | Chr1 | 2639069 | 2639317 | 248 | |
| Deletion | Chr8 | 3859022 | 3859035 | 13 | |
| Deletion | Chr7 | 4226611 | 4226617 | 6 | |
| Deletion | Chr7 | 4565801 | 4565811 | 10 | |
| Deletion | Chr5 | 5232158 | 5232169 | 11 | |
| Deletion | Chr5 | 5605780 | 5605789 | 9 | |
| Deletion | Chr3 | 5756551 | 5756557 | 6 | |
| Deletion | Chr2 | 7560675 | 7560687 | 12 | LOC_Os02g13910 |
| Deletion | Chr7 | 7710822 | 7710823 | 1 | |
| Deletion | Chr9 | 9033708 | 9033711 | 3 | LOC_Os09g15000 |
| Deletion | Chr5 | 9287519 | 9287520 | 1 | |
| Deletion | Chr5 | 10922063 | 10922068 | 5 | |
| Deletion | Chr5 | 12336130 | 12336132 | 2 | |
| Deletion | Chr7 | 12801767 | 12801771 | 4 | |
| Deletion | Chr10 | 15277844 | 15277851 | 7 | |
| Deletion | Chr10 | 15533065 | 15533074 | 9 | |
| Deletion | Chr5 | 17488260 | 17488261 | 1 | |
| Deletion | Chr10 | 18331952 | 18331959 | 7 | |
| Deletion | Chr6 | 20018728 | 20018737 | 9 | |
| Deletion | Chr4 | 25108393 | 25108394 | 1 | |
| Deletion | Chr8 | 25979334 | 25979341 | 7 | |
| Deletion | Chr8 | 25987251 | 25987253 | 2 | |
| Deletion | Chr8 | 26000906 | 26000923 | 17 | LOC_Os08g41160 |
| Deletion | Chr5 | 26153680 | 26153681 | 1 | LOC_Os05g44990 |
| Deletion | Chr4 | 26559480 | 26559522 | 42 | |
| Deletion | Chr1 | 27010202 | 27010214 | 12 | |
| Deletion | Chr7 | 27017001 | 27032000 | 14999 | 3 |
| Deletion | Chr6 | 29804227 | 29804233 | 6 | |
| Deletion | Chr3 | 31283663 | 31283685 | 22 | LOC_Os03g55010 |
| Deletion | Chr1 | 38493971 | 38493976 | 5 | LOC_Os01g66280 |
Insertions: 5
Inversions: 9
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 4082339 | 4082702 | LOC_Os06g08380 |
| Inversion | Chr8 | 5582929 | 5948786 | LOC_Os08g10244 |
| Inversion | Chr8 | 5582935 | 5753065 | LOC_Os08g09950 |
| Inversion | Chr10 | 11922292 | 11922458 | |
| Inversion | Chr10 | 21569605 | 22382694 | LOC_Os10g40260 |
| Inversion | Chr10 | 21569616 | 22382695 | 2 |
| Inversion | Chr4 | 23762764 | 23763013 | LOC_Os04g39900 |
| Inversion | Chr11 | 24120663 | 24121117 | LOC_Os11g40450 |
| Inversion | Chr4 | 30932193 | 30932639 | LOC_Os04g52074 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 21569616 | Chr5 | 22985125 | 2 |
| Translocation | Chr10 | 22382696 | Chr5 | 22984958 | LOC_Os05g39200 |
