Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN416-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN416-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 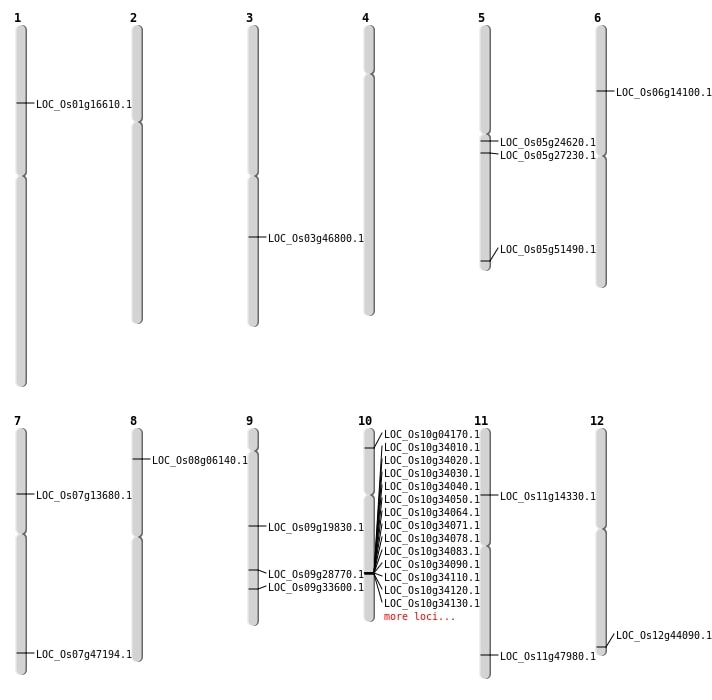
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14321212 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 34972919 | A-C | HET | INTRON | |
| SBS | Chr1 | 8936411 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr10 | 1938665 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g04170 |
| SBS | Chr10 | 2550 | T-G | HET | INTERGENIC | |
| SBS | Chr10 | 3424543 | A-T | HET | INTRON | |
| SBS | Chr11 | 19817463 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 27679693 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 8041730 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g14330 |
| SBS | Chr12 | 20826461 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 648899 | G-A | HET | INTRON | |
| SBS | Chr2 | 27088884 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 33676205 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 9394913 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 11142640 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 1629933 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 20828360 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 35207136 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr5 | 16805721 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 16805722 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 29525553 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g51490 |
| SBS | Chr5 | 986215 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 12883464 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 12905511 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 1968827 | A-G | HET | INTRON | |
| SBS | Chr6 | 27006873 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 4585575 | T-C | HET | INTRON | |
| SBS | Chr6 | 7865703 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g14100 |
| SBS | Chr7 | 28213714 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g47194 |
| SBS | Chr7 | 29126800 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 7853027 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g13680 |
| SBS | Chr8 | 17683255 | G-C | HET | INTRON | |
| SBS | Chr8 | 26360077 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 10082931 | G-A | HET | INTRON | |
| SBS | Chr9 | 12847017 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 15411625 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 16451659 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 21942907 | C-T | HET | INTRON | |
| SBS | Chr9 | 7480231 | T-C | HET | INTRON | |
| SBS | Chr9 | 9597736 | A-C | HET | INTERGENIC | |
| SBS | Chr9 | 9617490 | G-A | HET | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 3390765 | 3390770 | 5 | LOC_Os08g06140 |
| Deletion | Chr11 | 4772535 | 4772539 | 4 | |
| Deletion | Chr6 | 7735750 | 7735766 | 16 | |
| Deletion | Chr12 | 8727326 | 8727334 | 8 | |
| Deletion | Chr1 | 9430693 | 9430694 | 1 | LOC_Os01g16610 |
| Deletion | Chr3 | 9958895 | 9958898 | 3 | |
| Deletion | Chr5 | 10424439 | 10424459 | 20 | |
| Deletion | Chr9 | 11872607 | 11872962 | 355 | LOC_Os09g19830 |
| Deletion | Chr9 | 12518878 | 12518900 | 22 | |
| Deletion | Chr11 | 13136127 | 13136130 | 3 | |
| Deletion | Chr9 | 13221779 | 13221783 | 4 | |
| Deletion | Chr5 | 15830941 | 15830944 | 3 | LOC_Os05g27230 |
| Deletion | Chr5 | 16590824 | 16590827 | 3 | |
| Deletion | Chr8 | 17345643 | 17345667 | 24 | |
| Deletion | Chr10 | 18145001 | 18276000 | 131000 | 20 |
| Deletion | Chr11 | 18230984 | 18230985 | 1 | |
| Deletion | Chr1 | 20304720 | 20304729 | 9 | |
| Deletion | Chr3 | 21703729 | 21703740 | 11 | |
| Deletion | Chr1 | 25162004 | 25162007 | 3 | |
| Deletion | Chr5 | 27919624 | 27919625 | 1 | |
| Deletion | Chr3 | 28002556 | 28002560 | 4 | |
| Deletion | Chr6 | 29563720 | 29563721 | 1 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 2363633 | 2363633 | 1 | |
| Insertion | Chr12 | 10860859 | 10860859 | 1 | |
| Insertion | Chr2 | 945376 | 945377 | 2 | |
| Insertion | Chr3 | 26473136 | 26473136 | 1 | LOC_Os03g46800 |
| Insertion | Chr5 | 5300443 | 5300443 | 1 | |
| Insertion | Chr7 | 25253563 | 25253563 | 1 | |
| Insertion | Chr8 | 26815767 | 26815768 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 14223660 | 16178003 | LOC_Os05g24620 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 11872651 | Chr5 | 25341981 | LOC_Os09g19830 |
| Translocation | Chr9 | 11872953 | Chr5 | 25341977 | LOC_Os09g19830 |
| Translocation | Chr10 | 18276674 | Chr9 | 19828941 | LOC_Os09g33600 |
| Translocation | Chr10 | 18276683 | Chr4 | 16840329 | |
| Translocation | Chr10 | 18280408 | Chr4 | 16840294 | |
| Translocation | Chr11 | 25284672 | Chr9 | 17479450 | LOC_Os09g28770 |
| Translocation | Chr11 | 25284677 | Chr9 | 17495564 | |
| Translocation | Chr12 | 27344056 | Chr11 | 28939319 | 2 |
