Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN418-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN418-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 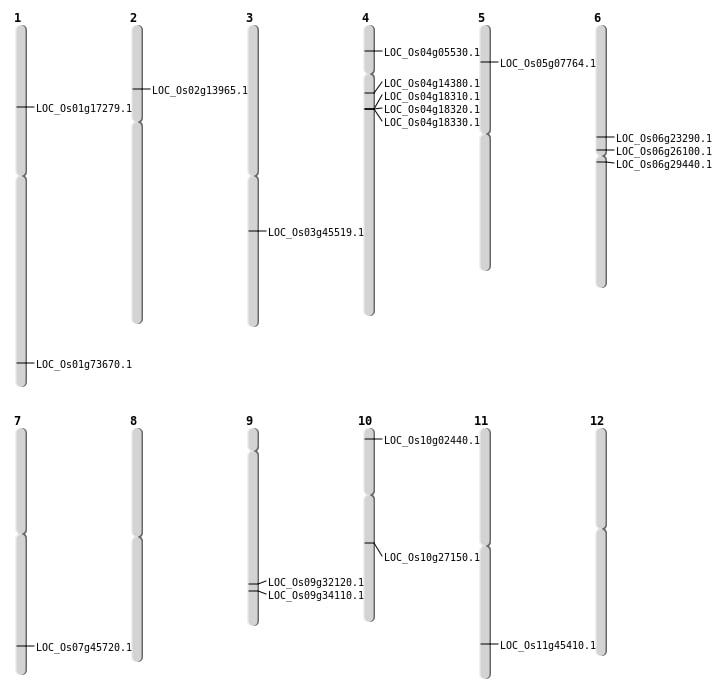
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 3497919 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 3628880 | G-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 36680017 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 14309284 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g27150 |
| SBS | Chr10 | 14451468 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 19559545 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 4934277 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 7935983 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 891275 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g02440 |
| SBS | Chr11 | 18266339 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 27496739 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g45410 |
| SBS | Chr11 | 8667933 | G-A | HET | INTRON | |
| SBS | Chr11 | 9402483 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 10317784 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 13089637 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 14455999 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 14456000 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 15822283 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 17199622 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 17444749 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 21131738 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 25744736 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 31894512 | G-A | HET | INTRON | |
| SBS | Chr4 | 1660776 | C-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 23774920 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 2797104 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g05530 |
| SBS | Chr4 | 28956080 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 32059465 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 15602732 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 15602733 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 15602735 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 19100569 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 8507500 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 13602806 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g23290 |
| SBS | Chr6 | 15273057 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g26100 |
| SBS | Chr6 | 27836106 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 28125924 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 16327858 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 18431318 | T-G | HET | UTR_3_PRIME | |
| SBS | Chr7 | 20809168 | G-T | HET | INTRON | |
| SBS | Chr7 | 26833401 | T-A | HOMO | INTRON | |
| SBS | Chr8 | 28115870 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 12500137 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 15352149 | T-A | HOMO | INTRON | |
| SBS | Chr9 | 20132497 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g34110 |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 1517496 | 1517517 | 21 | |
| Deletion | Chr9 | 3394458 | 3394459 | 1 | |
| Deletion | Chr5 | 4183378 | 4185986 | 2608 | LOC_Os05g07764 |
| Deletion | Chr2 | 4278651 | 4278652 | 1 | |
| Deletion | Chr9 | 5746380 | 5746382 | 2 | |
| Deletion | Chr5 | 7450633 | 7450637 | 4 | |
| Deletion | Chr2 | 7618793 | 7618806 | 13 | LOC_Os02g13965 |
| Deletion | Chr4 | 8050831 | 8050842 | 11 | LOC_Os04g14380 |
| Deletion | Chr1 | 9947516 | 9947519 | 3 | LOC_Os01g17279 |
| Deletion | Chr4 | 10108828 | 10114602 | 5774 | 3 |
| Deletion | Chr5 | 11067369 | 11067384 | 15 | |
| Deletion | Chr5 | 11895425 | 11895427 | 2 | |
| Deletion | Chr8 | 14009092 | 14009106 | 14 | |
| Deletion | Chr7 | 15017337 | 15017338 | 1 | |
| Deletion | Chr9 | 15314972 | 15314977 | 5 | |
| Deletion | Chr9 | 15955234 | 15955236 | 2 | |
| Deletion | Chr6 | 16864095 | 16864097 | 2 | LOC_Os06g29440 |
| Deletion | Chr6 | 20485520 | 20485524 | 4 | |
| Deletion | Chr4 | 26808045 | 26808052 | 7 | |
| Deletion | Chr11 | 27355058 | 27355065 | 7 | |
| Deletion | Chr1 | 42689356 | 42689369 | 13 | LOC_Os01g73670 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 4344845 | 4344845 | 1 | |
| Insertion | Chr10 | 23144919 | 23144919 | 1 | |
| Insertion | Chr12 | 20194941 | 20194942 | 2 | |
| Insertion | Chr12 | 25929890 | 25929893 | 4 |
No Inversion
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 19178791 | Chr3 | 25691877 | 2 |
| Translocation | Chr11 | 27838231 | Chr7 | 27296787 | LOC_Os07g45720 |
