Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4188-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4188-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 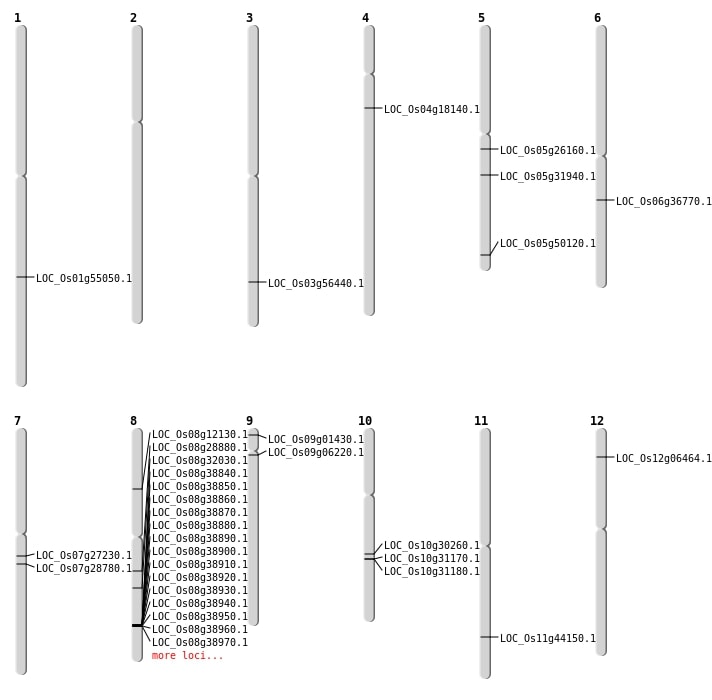
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24012716 | T-A | HET | ||
| SBS | Chr1 | 30565048 | C-T | HET | ||
| SBS | Chr1 | 3460010 | T-G | HET | ||
| SBS | Chr1 | 38343119 | G-A | HET | ||
| SBS | Chr10 | 11066372 | T-A | Homo | ||
| SBS | Chr10 | 15340092 | G-T | HET | ||
| SBS | Chr10 | 15721488 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g30260 |
| SBS | Chr10 | 18817619 | C-T | HET | ||
| SBS | Chr11 | 15483083 | A-G | HET | ||
| SBS | Chr11 | 21671760 | C-T | HET | ||
| SBS | Chr12 | 26091614 | T-A | HET | ||
| SBS | Chr2 | 11380350 | C-T | Homo | ||
| SBS | Chr2 | 11799978 | T-A | Homo | ||
| SBS | Chr2 | 26740546 | G-A | Homo | ||
| SBS | Chr3 | 32169790 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g56440 |
| SBS | Chr3 | 9643743 | T-A | HET | ||
| SBS | Chr4 | 10029014 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g18140 |
| SBS | Chr4 | 6106199 | G-C | HET | ||
| SBS | Chr5 | 15221847 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g26160 |
| SBS | Chr5 | 15221848 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g26160 |
| SBS | Chr5 | 25545484 | G-A | HET | ||
| SBS | Chr6 | 13843546 | T-A | HET | ||
| SBS | Chr6 | 14270898 | C-T | Homo | ||
| SBS | Chr6 | 20464777 | A-G | HET | ||
| SBS | Chr6 | 28741159 | C-T | HET | ||
| SBS | Chr7 | 16848389 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g28780 |
| SBS | Chr7 | 25475125 | G-A | HET | ||
| SBS | Chr8 | 17415149 | C-T | HET | ||
| SBS | Chr8 | 18376167 | C-T | HET | ||
| SBS | Chr8 | 21146435 | C-T | HET | ||
| SBS | Chr8 | 5092012 | A-G | HET | ||
| SBS | Chr9 | 12902467 | C-T | HET | ||
| SBS | Chr9 | 327373 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g01430 |
| SBS | Chr9 | 5502247 | A-G | HET | ||
| SBS | Chr9 | 9104138 | G-A | HET | ||
| SBS | Chr9 | 9437757 | A-G | HET |
Deletions: 33
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 274117 | 274120 | 3 | |
| Deletion | Chr12 | 774057 | 774058 | 1 | |
| Deletion | Chr1 | 4974770 | 4974773 | 3 | |
| Deletion | Chr8 | 5538703 | 5538705 | 2 | |
| Deletion | Chr7 | 6202485 | 6202486 | 1 | |
| Deletion | Chr4 | 7002359 | 7002369 | 10 | |
| Deletion | Chr9 | 7060358 | 7060364 | 6 | |
| Deletion | Chr2 | 8105383 | 8105389 | 6 | |
| Deletion | Chr2 | 10099465 | 10099483 | 18 | |
| Deletion | Chr1 | 10649201 | 10649202 | 1 | |
| Deletion | Chr2 | 12327537 | 12327540 | 3 | |
| Deletion | Chr4 | 12758441 | 12758442 | 1 | |
| Deletion | Chr6 | 15618096 | 15618115 | 19 | |
| Deletion | Chr2 | 16215555 | 16215564 | 9 | |
| Deletion | Chr10 | 16316001 | 16349000 | 32999 | 2 |
| Deletion | Chr5 | 16616535 | 16616547 | 12 | |
| Deletion | Chr7 | 18596359 | 18596367 | 8 | |
| Deletion | Chr5 | 18604406 | 18604407 | 1 | LOC_Os05g31940 |
| Deletion | Chr7 | 18949065 | 18949074 | 9 | |
| Deletion | Chr4 | 19166379 | 19166380 | 1 | |
| Deletion | Chr11 | 19209118 | 19209135 | 17 | |
| Deletion | Chr8 | 19652090 | 19652091 | 1 | |
| Deletion | Chr8 | 19869248 | 19869256 | 8 | LOC_Os08g32030 |
| Deletion | Chr3 | 20845457 | 20845474 | 17 | |
| Deletion | Chr12 | 21944858 | 21944865 | 7 | |
| Deletion | Chr5 | 23691837 | 23691840 | 3 | |
| Deletion | Chr8 | 24546001 | 24708000 | 161999 | 26 |
| Deletion | Chr11 | 26654706 | 26654709 | 3 | LOC_Os11g44150 |
| Deletion | Chr5 | 27408986 | 27408987 | 1 | |
| Deletion | Chr3 | 29453179 | 29453190 | 11 | |
| Deletion | Chr5 | 29702442 | 29702444 | 2 | |
| Deletion | Chr3 | 31744087 | 31744090 | 3 | |
| Deletion | Chr4 | 32084074 | 32084075 | 1 |
Insertions: 8
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 9766031 | 9838531 | |
| Inversion | Chr10 | 15836662 | 16345555 | |
| Inversion | Chr8 | 17665662 | 17991617 | LOC_Os08g28880 |
| Inversion | Chr8 | 17668306 | 17991620 | |
| Inversion | Chr1 | 31654145 | 31859144 | LOC_Os01g55050 |
| Inversion | Chr1 | 31654149 | 31859156 | LOC_Os01g55050 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 2908289 | Chr8 | 10553493 | LOC_Os09g06220 |
| Translocation | Chr9 | 2909810 | Chr8 | 10553480 | LOC_Os09g06220 |
| Translocation | Chr12 | 3105411 | Chr3 | 25259154 | LOC_Os12g06464 |
| Translocation | Chr5 | 4616136 | Chr3 | 5945872 | |
| Translocation | Chr8 | 7146011 | Chr7 | 15823038 | 2 |
| Translocation | Chr10 | 12580401 | Chr8 | 14922478 | |
| Translocation | Chr6 | 21657632 | Chr4 | 17922848 | LOC_Os06g36770 |
| Translocation | Chr5 | 28724711 | Chr3 | 9391687 | LOC_Os05g50120 |
