Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4189-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4189-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 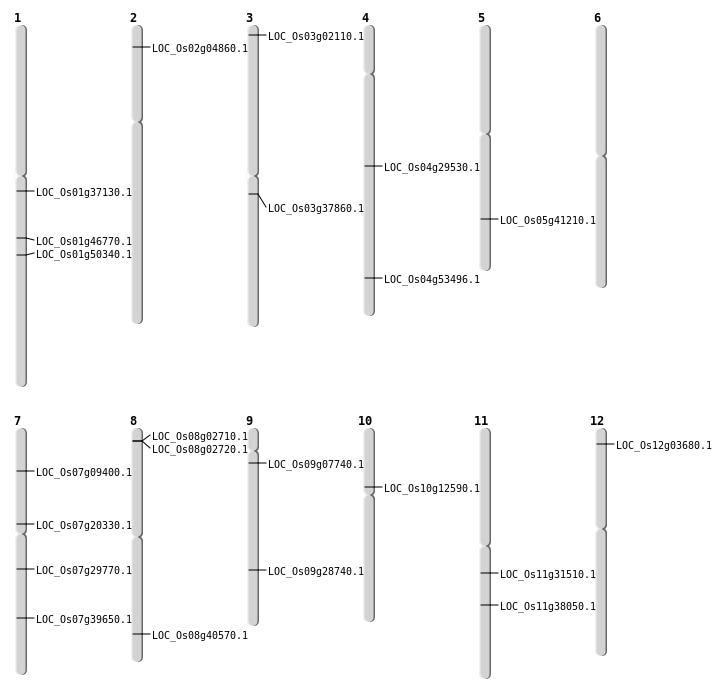
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13695842 | G-A | HET | ||
| SBS | Chr1 | 15793169 | T-G | Homo | ||
| SBS | Chr1 | 29005993 | G-A | HET | ||
| SBS | Chr1 | 29542577 | G-A | HET | ||
| SBS | Chr1 | 34919998 | G-C | HET | ||
| SBS | Chr1 | 41325450 | T-C | HET | ||
| SBS | Chr11 | 18417572 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g31510 |
| SBS | Chr11 | 18873839 | G-A | HET | ||
| SBS | Chr11 | 19972079 | A-C | HET | ||
| SBS | Chr11 | 23446167 | G-C | Homo | ||
| SBS | Chr11 | 5733968 | T-C | HET | ||
| SBS | Chr12 | 13370149 | T-C | Homo | ||
| SBS | Chr2 | 11285563 | G-A | Homo | ||
| SBS | Chr2 | 17041689 | C-T | Homo | ||
| SBS | Chr2 | 6387602 | G-C | HET | ||
| SBS | Chr2 | 6579241 | T-C | Homo | ||
| SBS | Chr3 | 32893619 | G-C | HET | ||
| SBS | Chr3 | 679989 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g02110 |
| SBS | Chr4 | 10347778 | A-T | HET | ||
| SBS | Chr4 | 16067591 | A-T | Homo | ||
| SBS | Chr4 | 30754487 | A-T | HET | ||
| SBS | Chr4 | 30754488 | A-T | HET | ||
| SBS | Chr4 | 31856452 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g53496 |
| SBS | Chr4 | 31856453 | G-A | HET | ||
| SBS | Chr4 | 31856454 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g53496 |
| SBS | Chr5 | 13657156 | G-A | Homo | ||
| SBS | Chr5 | 15368861 | A-T | Homo | ||
| SBS | Chr5 | 7556626 | T-G | Homo | ||
| SBS | Chr5 | 7556627 | T-C | Homo | ||
| SBS | Chr5 | 8560094 | T-G | HET | ||
| SBS | Chr5 | 9276527 | T-A | HET | ||
| SBS | Chr6 | 27545017 | C-T | Homo | ||
| SBS | Chr7 | 11730358 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g20330 |
| SBS | Chr7 | 26744823 | T-C | Homo | ||
| SBS | Chr8 | 25684591 | G-A | HET | STOP_GAINED | LOC_Os08g40570 |
| SBS | Chr9 | 4107808 | C-G | HET |
Deletions: 11
Insertions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 26670083 | 26670087 | 5 | LOC_Os01g46770 |
| Insertion | Chr1 | 34047786 | 34047786 | 1 | |
| Insertion | Chr10 | 1412742 | 1412742 | 1 | |
| Insertion | Chr11 | 21129520 | 21129533 | 14 | |
| Insertion | Chr11 | 22104942 | 22104953 | 12 | |
| Insertion | Chr11 | 23637084 | 23637084 | 1 | |
| Insertion | Chr11 | 27010692 | 27010692 | 1 | |
| Insertion | Chr12 | 15721069 | 15721074 | 6 | |
| Insertion | Chr5 | 26651070 | 26651071 | 2 | |
| Insertion | Chr6 | 27127559 | 27127560 | 2 | |
| Insertion | Chr7 | 12594231 | 12594231 | 1 | |
| Insertion | Chr7 | 19025978 | 19025985 | 8 | |
| Insertion | Chr7 | 24530941 | 24530941 | 1 | |
| Insertion | Chr7 | 26105674 | 26105674 | 1 | |
| Insertion | Chr9 | 3909775 | 3909776 | 2 | LOC_Os09g07740 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 6555273 | 7011632 | LOC_Os10g12590 |
| Inversion | Chr10 | 6555274 | 7011635 | LOC_Os10g12590 |
| Inversion | Chr9 | 17447768 | 17469262 | LOC_Os09g28740 |
| Inversion | Chr9 | 17447779 | 17469272 | LOC_Os09g28740 |
Translocations: 18
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 1131575 | Chr2 | 2245050 | 2 |
| Translocation | Chr8 | 1139628 | Chr2 | 3825319 | LOC_Os08g02720 |
| Translocation | Chr12 | 1482576 | Chr2 | 9635657 | LOC_Os12g03680 |
| Translocation | Chr7 | 4957699 | Chr1 | 28896027 | 2 |
| Translocation | Chr7 | 4957960 | Chr1 | 28896026 | 2 |
| Translocation | Chr10 | 5623929 | Chr6 | 8458373 | |
| Translocation | Chr8 | 9700385 | Chr5 | 22608731 | |
| Translocation | Chr5 | 11149282 | Chr4 | 17545687 | LOC_Os04g29530 |
| Translocation | Chr4 | 13338103 | Chr2 | 2245094 | LOC_Os02g04860 |
| Translocation | Chr4 | 13339770 | Chr3 | 21021250 | LOC_Os03g37860 |
| Translocation | Chr6 | 16399197 | Chr2 | 26377474 | |
| Translocation | Chr7 | 17505067 | Chr5 | 24133852 | 2 |
| Translocation | Chr7 | 17505327 | Chr5 | 24133851 | 2 |
| Translocation | Chr5 | 17627891 | Chr1 | 42980730 | |
| Translocation | Chr11 | 20079919 | Chr6 | 19269355 | |
| Translocation | Chr11 | 22570659 | Chr7 | 4765763 | LOC_Os11g38050 |
| Translocation | Chr7 | 23767442 | Chr1 | 20739018 | 2 |
| Translocation | Chr11 | 26139354 | Chr9 | 13620998 |
