Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4234-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4234-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 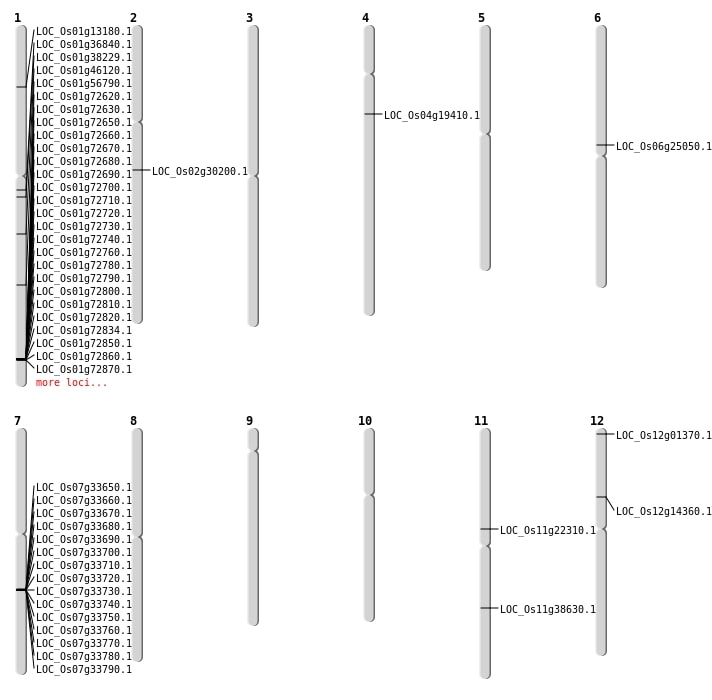
|
| Variant Information |
|---|
Single base substitutions: 61
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15219485 | G-A | HET | ||
| SBS | Chr1 | 22762439 | G-C | HET | ||
| SBS | Chr1 | 24582620 | T-A | HET | ||
| SBS | Chr1 | 24734267 | C-T | HET | ||
| SBS | Chr1 | 27795284 | C-T | Homo | ||
| SBS | Chr1 | 32646318 | T-A | Homo | ||
| SBS | Chr1 | 32772105 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g56790 |
| SBS | Chr1 | 7346312 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g13180 |
| SBS | Chr1 | 9234106 | C-T | HET | ||
| SBS | Chr10 | 2940067 | A-G | Homo | ||
| SBS | Chr10 | 5384461 | G-A | HET | ||
| SBS | Chr11 | 16054912 | G-A | HET | ||
| SBS | Chr11 | 18086187 | A-G | HET | ||
| SBS | Chr11 | 21060465 | T-A | HET | ||
| SBS | Chr11 | 25892805 | A-G | Homo | ||
| SBS | Chr11 | 26645837 | G-A | Homo | ||
| SBS | Chr11 | 5341922 | G-A | Homo | ||
| SBS | Chr12 | 12504911 | T-A | HET | ||
| SBS | Chr12 | 14638366 | G-A | HET | ||
| SBS | Chr12 | 26404868 | T-C | HET | ||
| SBS | Chr12 | 5650994 | G-A | HET | ||
| SBS | Chr12 | 8181674 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g14360 |
| SBS | Chr2 | 10533999 | G-A | HET | ||
| SBS | Chr2 | 15056048 | C-T | HET | ||
| SBS | Chr2 | 17266880 | C-T | HET | ||
| SBS | Chr2 | 1889435 | A-G | HET | ||
| SBS | Chr2 | 24891965 | G-A | HET | ||
| SBS | Chr2 | 30539923 | A-G | HET | ||
| SBS | Chr2 | 33182473 | G-C | HET | ||
| SBS | Chr2 | 35921719 | G-C | HET | ||
| SBS | Chr3 | 15174038 | C-T | Homo | ||
| SBS | Chr3 | 2776482 | A-T | HET | ||
| SBS | Chr3 | 31778911 | A-C | Homo | ||
| SBS | Chr3 | 31994016 | G-T | HET | ||
| SBS | Chr3 | 3753631 | A-G | HET | ||
| SBS | Chr3 | 9529766 | T-C | HET | ||
| SBS | Chr4 | 10815079 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g19410 |
| SBS | Chr4 | 11368023 | T-A | HET | ||
| SBS | Chr4 | 12418383 | A-C | HET | ||
| SBS | Chr4 | 19711691 | T-C | HET | ||
| SBS | Chr4 | 19841436 | G-A | HET | ||
| SBS | Chr4 | 34433053 | G-A | HET | ||
| SBS | Chr5 | 21438407 | C-G | HET | ||
| SBS | Chr5 | 29418044 | G-A | HET | ||
| SBS | Chr5 | 8405741 | C-T | HET | ||
| SBS | Chr6 | 14658488 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g25050 |
| SBS | Chr6 | 16121662 | G-A | HET | ||
| SBS | Chr6 | 16685113 | G-A | HET | ||
| SBS | Chr6 | 4912842 | G-T | HET | ||
| SBS | Chr7 | 18998620 | C-T | Homo | ||
| SBS | Chr7 | 25312166 | A-T | Homo | ||
| SBS | Chr7 | 313913 | C-G | HET | ||
| SBS | Chr8 | 12851362 | G-A | HET | ||
| SBS | Chr8 | 16960354 | T-C | HET | ||
| SBS | Chr8 | 22537506 | A-T | HET | ||
| SBS | Chr8 | 23136925 | G-A | HET | ||
| SBS | Chr8 | 23796922 | T-C | HET | ||
| SBS | Chr8 | 4295588 | G-C | HET | ||
| SBS | Chr9 | 14780712 | G-A | HET | ||
| SBS | Chr9 | 1708093 | C-T | Homo | ||
| SBS | Chr9 | 3417586 | G-A | HET |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 855933 | 855935 | 2 | |
| Deletion | Chr5 | 1691821 | 1691823 | 2 | |
| Deletion | Chr1 | 1797537 | 1797555 | 18 | |
| Deletion | Chr7 | 3554709 | 3554726 | 17 | |
| Deletion | Chr10 | 5482208 | 5482209 | 1 | |
| Deletion | Chr2 | 9119265 | 9119295 | 30 | |
| Deletion | Chr11 | 12792500 | 12792517 | 17 | LOC_Os11g22310 |
| Deletion | Chr10 | 19520467 | 19520480 | 13 | |
| Deletion | Chr7 | 20099001 | 20231000 | 131999 | 15 |
| Deletion | Chr11 | 21916012 | 21916018 | 6 | |
| Deletion | Chr12 | 22040816 | 22040833 | 17 | |
| Deletion | Chr11 | 22901036 | 22901042 | 6 | LOC_Os11g38630 |
| Deletion | Chr11 | 23192682 | 23192683 | 1 | |
| Deletion | Chr3 | 23504897 | 23504903 | 6 | |
| Deletion | Chr3 | 25036357 | 25036361 | 4 | |
| Deletion | Chr1 | 26222812 | 26222814 | 2 | LOC_Os01g46120 |
| Deletion | Chr1 | 26501474 | 26501476 | 2 | |
| Deletion | Chr11 | 26759330 | 26759332 | 2 | |
| Deletion | Chr7 | 26828542 | 26828543 | 1 | |
| Deletion | Chr1 | 42131001 | 42420000 | 288999 | 47 |
Insertions: 7
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 8179729 | 8180140 | |
| Inversion | Chr1 | 20497701 | 21428922 | 2 |
| Inversion | Chr1 | 20497706 | 21428960 | 2 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 229431 | Chr11 | 204190 | LOC_Os12g01370 |
| Translocation | Chr10 | 5457160 | Chr2 | 14657860 | |
| Translocation | Chr3 | 8825037 | Chr2 | 17955708 | LOC_Os02g30200 |
| Translocation | Chr9 | 14951257 | Chr1 | 37409863 | |
| Translocation | Chr12 | 18630149 | Chr7 | 6891519 | |
| Translocation | Chr7 | 20101660 | Chr1 | 22096382 | |
| Translocation | Chr7 | 20230651 | Chr1 | 22096367 | |
| Translocation | Chr11 | 25027911 | Chr8 | 27374348 |
