Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4272-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4272-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 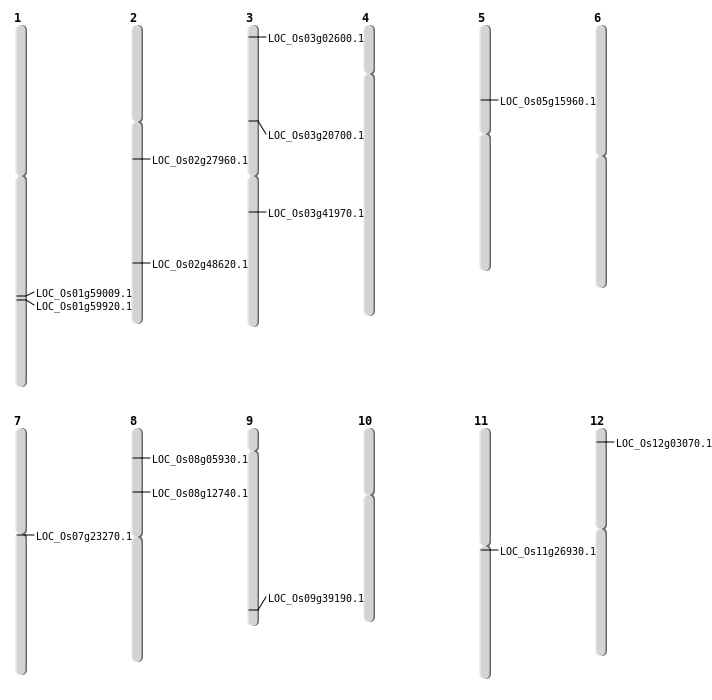
|
| Variant Information |
|---|
Single base substitutions: 52
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 34114152 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g59009 |
| SBS | Chr1 | 3730190 | C-T | HET | ||
| SBS | Chr1 | 38163154 | G-A | Homo | ||
| SBS | Chr1 | 4099654 | G-A | HET | ||
| SBS | Chr10 | 2421564 | G-A | HET | ||
| SBS | Chr10 | 8617805 | C-T | HET | ||
| SBS | Chr11 | 15485951 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g26930 |
| SBS | Chr11 | 7578561 | C-A | HET | ||
| SBS | Chr11 | 9956582 | T-A | HET | ||
| SBS | Chr11 | 9956583 | T-A | HET | ||
| SBS | Chr12 | 16486103 | A-T | HET | ||
| SBS | Chr12 | 24783555 | G-A | HET | ||
| SBS | Chr12 | 5730047 | A-T | Homo | ||
| SBS | Chr12 | 8771249 | T-C | Homo | ||
| SBS | Chr2 | 12188643 | T-A | HET | ||
| SBS | Chr2 | 24239581 | G-A | HET | ||
| SBS | Chr2 | 30188885 | A-G | HET | ||
| SBS | Chr2 | 30236095 | A-C | HET | ||
| SBS | Chr2 | 31113455 | C-T | HET | ||
| SBS | Chr2 | 32201101 | A-G | HET | ||
| SBS | Chr2 | 5035683 | C-G | HET | ||
| SBS | Chr2 | 9214048 | G-A | HET | ||
| SBS | Chr3 | 10401417 | C-A | HET | ||
| SBS | Chr3 | 12472619 | G-C | HET | ||
| SBS | Chr3 | 20317530 | C-A | Homo | ||
| SBS | Chr3 | 20317531 | T-C | Homo | ||
| SBS | Chr3 | 24462067 | C-T | Homo | ||
| SBS | Chr3 | 25503142 | A-G | Homo | ||
| SBS | Chr3 | 26836684 | C-T | HET | ||
| SBS | Chr3 | 2934516 | C-A | HET | ||
| SBS | Chr3 | 4022225 | G-T | HET | ||
| SBS | Chr3 | 4022229 | T-A | HET | ||
| SBS | Chr4 | 15423995 | G-A | HET | ||
| SBS | Chr4 | 15423997 | G-A | HET | ||
| SBS | Chr4 | 197170 | A-T | HET | ||
| SBS | Chr4 | 197173 | A-G | HET | ||
| SBS | Chr4 | 8062226 | A-G | HET | ||
| SBS | Chr5 | 15370221 | C-T | HET | ||
| SBS | Chr6 | 11995896 | A-T | Homo | ||
| SBS | Chr6 | 27570998 | C-T | HET | ||
| SBS | Chr6 | 835149 | G-A | HET | ||
| SBS | Chr7 | 13118716 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g23270 |
| SBS | Chr7 | 13360927 | C-T | Homo | ||
| SBS | Chr7 | 19224789 | T-C | HET | ||
| SBS | Chr7 | 20253924 | C-G | Homo | ||
| SBS | Chr8 | 10759902 | G-A | HET | ||
| SBS | Chr8 | 3200510 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g05930 |
| SBS | Chr8 | 6451475 | G-A | HET | ||
| SBS | Chr9 | 16927529 | T-C | HET | ||
| SBS | Chr9 | 17271970 | G-C | HET | ||
| SBS | Chr9 | 20652700 | G-A | HET | ||
| SBS | Chr9 | 5208839 | C-T | HET |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 1157049 | 1157056 | 7 | LOC_Os12g03070 |
| Deletion | Chr8 | 1684837 | 1684838 | 1 | |
| Deletion | Chr10 | 3649972 | 3649978 | 6 | |
| Deletion | Chr12 | 3753119 | 3753120 | 1 | |
| Deletion | Chr8 | 4945818 | 4945833 | 15 | |
| Deletion | Chr3 | 6783277 | 6783290 | 13 | |
| Deletion | Chr8 | 7550678 | 7550687 | 9 | LOC_Os08g12740 |
| Deletion | Chr12 | 8867300 | 8867301 | 1 | |
| Deletion | Chr5 | 9012651 | 9012656 | 5 | LOC_Os05g15960 |
| Deletion | Chr4 | 9295803 | 9295819 | 16 | |
| Deletion | Chr12 | 11208193 | 11208194 | 1 | |
| Deletion | Chr3 | 11713600 | 11713609 | 9 | LOC_Os03g20700 |
| Deletion | Chr2 | 13116534 | 13116535 | 1 | |
| Deletion | Chr4 | 18283548 | 18283549 | 1 | |
| Deletion | Chr3 | 20693537 | 20693549 | 12 | |
| Deletion | Chr2 | 21524992 | 21524994 | 2 | |
| Deletion | Chr3 | 23321236 | 23321240 | 4 | LOC_Os03g41970 |
| Deletion | Chr12 | 23878319 | 23878328 | 9 | |
| Deletion | Chr8 | 24311132 | 24311133 | 1 | |
| Deletion | Chr5 | 25521244 | 25521255 | 11 | |
| Deletion | Chr12 | 26045444 | 26045449 | 5 | |
| Deletion | Chr12 | 26618097 | 26618098 | 1 | |
| Deletion | Chr6 | 28442383 | 28442387 | 4 | |
| Deletion | Chr5 | 28849867 | 28849869 | 2 | |
| Deletion | Chr2 | 29765541 | 29765547 | 6 | LOC_Os02g48620 |
| Deletion | Chr1 | 30301099 | 30301109 | 10 | |
| Deletion | Chr1 | 34647811 | 34647815 | 4 | LOC_Os01g59920 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 13408578 | 13408623 | 46 | |
| Insertion | Chr10 | 2694374 | 2694535 | 162 | 2 |
| Insertion | Chr10 | 2694374 | 2694457 | 84 | 2 |
| Insertion | Chr3 | 26960155 | 26960156 | 2 | |
| Insertion | Chr3 | 30631040 | 30631041 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 16330508 | 16330754 | |
| Inversion | Chr6 | 27554382 | 27592518 | |
| Inversion | Chr6 | 27554389 | 27592522 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 4253378 | Chr3 | 961083 | LOC_Os03g02600 |
| Translocation | Chr10 | 4253569 | Chr3 | 961092 | LOC_Os03g02600 |
| Translocation | Chr9 | 22509951 | Chr2 | 16553864 | LOC_Os09g39190 |
| Translocation | Chr9 | 22509954 | Chr2 | 16552341 | 2 |
| Translocation | Chr5 | 25574137 | Chr4 | 24171125 |
