Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN428-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN428-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 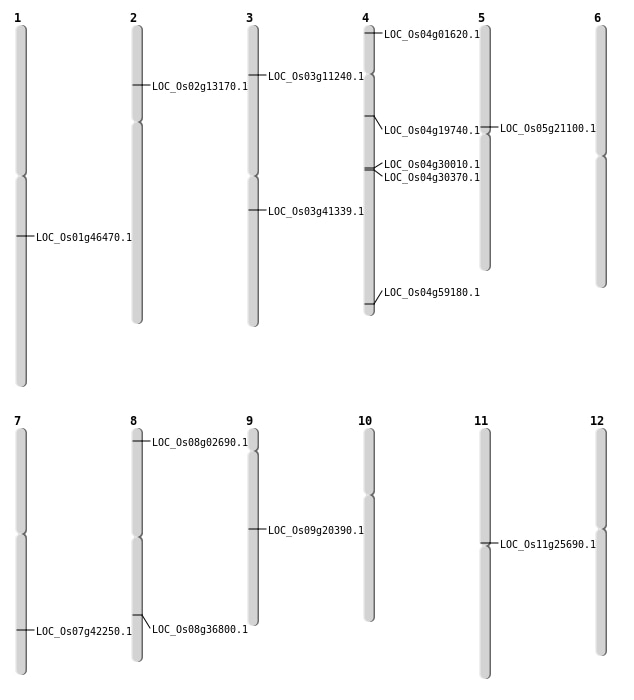
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13358277 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 23592423 | C-T | HET | INTRON | |
| SBS | Chr10 | 11852717 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 14712251 | C-G | HET | INTRON | |
| SBS | Chr10 | 2054499 | G-T | HET | INTRON | |
| SBS | Chr10 | 5705650 | A-G | HET | INTRON | |
| SBS | Chr10 | 7364121 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 8142093 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 10387381 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 14656173 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g25690 |
| SBS | Chr11 | 16059261 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 20921260 | C-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 4808469 | G-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 14192153 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 19493815 | C-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 6630619 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 6630628 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 11868403 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 15044899 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 15816535 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 24637602 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 26149653 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 33250445 | A-G | HET | INTRON | |
| SBS | Chr4 | 35200829 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g59180 |
| SBS | Chr5 | 12405915 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g21100 |
| SBS | Chr6 | 1027341 | G-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 11008668 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 30997112 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 19699419 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 23211636 | C-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 26170635 | A-T | HET | INTRON | |
| SBS | Chr7 | 6481069 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 14182835 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 1433899 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 16627322 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 2575790 | G-A | HET | INTRON | |
| SBS | Chr9 | 9031731 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 9031738 | C-T | HET | INTERGENIC |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 422623 | 422629 | 6 | LOC_Os04g01620 |
| Deletion | Chr2 | 491313 | 491334 | 21 | |
| Deletion | Chr8 | 1121407 | 1121411 | 4 | LOC_Os08g02690 |
| Deletion | Chr6 | 1192464 | 1192466 | 2 | |
| Deletion | Chr10 | 5219287 | 5219291 | 4 | |
| Deletion | Chr3 | 5781451 | 5782277 | 826 | LOC_Os03g11240 |
| Deletion | Chr2 | 6189654 | 6189656 | 2 | |
| Deletion | Chr12 | 6859950 | 6859951 | 1 | |
| Deletion | Chr9 | 7159892 | 7159897 | 5 | |
| Deletion | Chr5 | 8968421 | 8968434 | 13 | |
| Deletion | Chr4 | 11029285 | 11029294 | 9 | LOC_Os04g19740 |
| Deletion | Chr9 | 12253110 | 12253113 | 3 | LOC_Os09g20390 |
| Deletion | Chr12 | 12715861 | 12715862 | 1 | |
| Deletion | Chr10 | 14697829 | 14697838 | 9 | |
| Deletion | Chr9 | 16043995 | 16044000 | 5 | |
| Deletion | Chr9 | 16548207 | 16548208 | 1 | |
| Deletion | Chr5 | 17472181 | 17472182 | 1 | |
| Deletion | Chr9 | 17644829 | 17644836 | 7 | |
| Deletion | Chr11 | 19403217 | 19403218 | 1 | |
| Deletion | Chr9 | 19601922 | 19601936 | 14 | |
| Deletion | Chr3 | 22984824 | 22985481 | 657 | LOC_Os03g41339 |
| Deletion | Chr5 | 23208382 | 23208383 | 1 | |
| Deletion | Chr8 | 23251303 | 23251307 | 4 | LOC_Os08g36800 |
| Deletion | Chr7 | 23387657 | 23387666 | 9 | |
| Deletion | Chr3 | 23552282 | 23552318 | 36 | |
| Deletion | Chr3 | 24390321 | 24390323 | 2 | |
| Deletion | Chr3 | 24546616 | 24546618 | 2 | |
| Deletion | Chr1 | 26434144 | 26434145 | 1 | LOC_Os01g46470 |
| Deletion | Chr11 | 28234919 | 28234920 | 1 | |
| Deletion | Chr3 | 32648348 | 32648356 | 8 | |
| Deletion | Chr2 | 32781237 | 32781246 | 9 | |
| Deletion | Chr3 | 34550489 | 34550493 | 4 | |
| Deletion | Chr4 | 35356806 | 35356809 | 3 | |
| Deletion | Chr1 | 42687683 | 42687693 | 10 |
Insertions: 5
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 5874016 | 6749964 | |
| Inversion | Chr2 | 6195499 | 6749889 | |
| Inversion | Chr2 | 6841314 | 7023466 | LOC_Os02g13170 |
| Inversion | Chr2 | 6841329 | 7023462 | LOC_Os02g13170 |
| Inversion | Chr4 | 17906985 | 18125369 | 2 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 166474 | Chr3 | 1013848 | |
| Translocation | Chr9 | 9030562 | Chr3 | 5781464 | LOC_Os03g11240 |
| Translocation | Chr8 | 9498456 | Chr7 | 25287862 | LOC_Os07g42250 |
| Translocation | Chr9 | 19096342 | Chr8 | 21862201 | |
| Translocation | Chr9 | 19097131 | Chr8 | 21862189 |
