Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN430-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN430-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 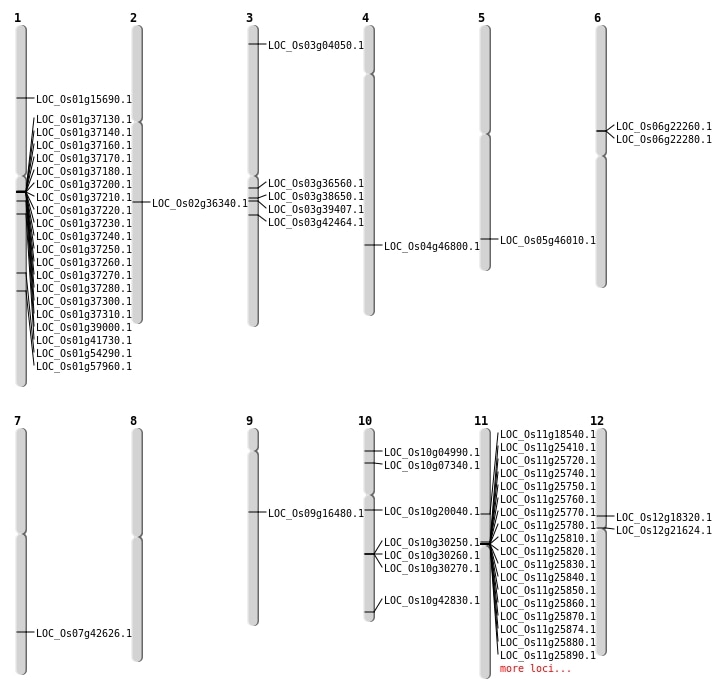
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21926167 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g39000 |
| SBS | Chr1 | 31235620 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g54290 |
| SBS | Chr10 | 100241 | G-C | HET | INTERGENIC | |
| SBS | Chr10 | 16460233 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 21457844 | T-C | HET | INTRON | |
| SBS | Chr10 | 2430615 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g04990 |
| SBS | Chr11 | 25475478 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g42300 |
| SBS | Chr11 | 25475479 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 10575284 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g18320 |
| SBS | Chr12 | 10575289 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g18320 |
| SBS | Chr12 | 12153482 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g21624 |
| SBS | Chr12 | 12153483 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g21624 |
| SBS | Chr12 | 18212698 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 24758534 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 24962269 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 13986527 | A-G | HOMO | INTRON | |
| SBS | Chr2 | 13986528 | G-A | HOMO | INTRON | |
| SBS | Chr2 | 15458359 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 15700760 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 16516010 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 32917087 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr2 | 33430160 | A-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 12967635 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 26550315 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 10062839 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 21017815 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 24415888 | G-A | HOMO | INTRON | |
| SBS | Chr5 | 26671766 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g46010 |
| SBS | Chr6 | 14339922 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 15111754 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 28126468 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 21094446 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 25521580 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g42626 |
| SBS | Chr8 | 11489217 | T-G | HET | INTRON | |
| SBS | Chr8 | 13911424 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 25259276 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 25511045 | T-C | HET | INTERGENIC |
Deletions: 35
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 440877 | 440882 | 5 | |
| Deletion | Chr2 | 763952 | 763966 | 14 | |
| Deletion | Chr2 | 1364036 | 1364061 | 25 | |
| Deletion | Chr3 | 1856520 | 1856522 | 2 | LOC_Os03g04050 |
| Deletion | Chr9 | 3194978 | 3195155 | 177 | |
| Deletion | Chr10 | 3869582 | 3869599 | 17 | LOC_Os10g07340 |
| Deletion | Chr8 | 6057490 | 6057497 | 7 | |
| Deletion | Chr1 | 8826806 | 8826812 | 6 | LOC_Os01g15690 |
| Deletion | Chr12 | 9189526 | 9189538 | 12 | |
| Deletion | Chr10 | 10056001 | 10062000 | 6000 | LOC_Os10g20040 |
| Deletion | Chr9 | 10097111 | 10097126 | 15 | LOC_Os09g16480 |
| Deletion | Chr12 | 10259961 | 10259982 | 21 | |
| Deletion | Chr4 | 10839700 | 10839701 | 1 | |
| Deletion | Chr5 | 12228801 | 12228813 | 12 | |
| Deletion | Chr6 | 12919587 | 12930552 | 10965 | 2 |
| Deletion | Chr11 | 14674355 | 14874960 | 200605 | 27 |
| Deletion | Chr10 | 15708001 | 15727000 | 19000 | 3 |
| Deletion | Chr11 | 17394001 | 17488000 | 94000 | 14 |
| Deletion | Chr8 | 20142382 | 20142383 | 1 | |
| Deletion | Chr1 | 20726001 | 20827000 | 101000 | 16 |
| Deletion | Chr5 | 21424237 | 21424239 | 2 | |
| Deletion | Chr3 | 21460586 | 21460590 | 4 | LOC_Os03g38650 |
| Deletion | Chr3 | 21903750 | 21903751 | 1 | LOC_Os03g39407 |
| Deletion | Chr8 | 21905176 | 21905181 | 5 | |
| Deletion | Chr1 | 21926150 | 21926153 | 3 | LOC_Os01g39000 |
| Deletion | Chr2 | 21934806 | 21934813 | 7 | LOC_Os02g36340 |
| Deletion | Chr1 | 22236815 | 22236816 | 1 | |
| Deletion | Chr10 | 23098367 | 23098379 | 12 | LOC_Os10g42830 |
| Deletion | Chr1 | 23626119 | 23626120 | 1 | LOC_Os01g41730 |
| Deletion | Chr3 | 23627668 | 23627915 | 247 | LOC_Os03g42464 |
| Deletion | Chr11 | 25151571 | 25151583 | 12 | |
| Deletion | Chr11 | 25544811 | 25544812 | 1 | |
| Deletion | Chr1 | 28108216 | 28108217 | 1 | |
| Deletion | Chr2 | 34078381 | 34078394 | 13 | |
| Deletion | Chr4 | 34169573 | 34169581 | 8 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr5 | 5970091 | 5970092 | 2 | |
| Insertion | Chr6 | 25813057 | 25813060 | 4 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 7229235 | 33506512 | LOC_Os01g57960 |
| Inversion | Chr2 | 14153703 | 15172623 | |
| Inversion | Chr11 | 14488126 | 14874960 | 2 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 134551 | Chr4 | 27738423 | LOC_Os04g46800 |
| Translocation | Chr11 | 10449195 | Chr6 | 17476342 | LOC_Os11g18540 |
| Translocation | Chr12 | 13397062 | Chr9 | 7173807 | |
| Translocation | Chr9 | 16353861 | Chr2 | 18616565 | |
| Translocation | Chr11 | 20393605 | Chr7 | 18351882 | |
| Translocation | Chr6 | 23038834 | Chr3 | 20268687 | LOC_Os03g36560 |
