Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4325-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4325-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 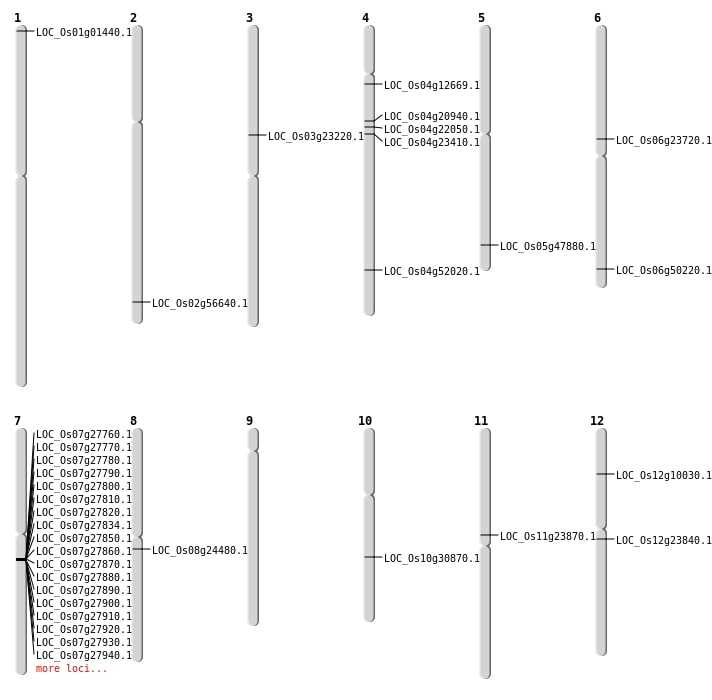
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr10 | 5665308 | A-T | HET | ||
| SBS | Chr11 | 1044288 | C-T | Homo | ||
| SBS | Chr11 | 13516990 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g23870 |
| SBS | Chr11 | 15001247 | A-G | HET | ||
| SBS | Chr11 | 2391077 | G-A | Homo | ||
| SBS | Chr11 | 3824162 | T-C | Homo | ||
| SBS | Chr12 | 13556044 | C-A | HET | STOP_GAINED | LOC_Os12g23840 |
| SBS | Chr12 | 15573320 | C-T | Homo | ||
| SBS | Chr12 | 19145987 | A-G | HET | ||
| SBS | Chr12 | 26206069 | A-T | HET | ||
| SBS | Chr12 | 7544149 | G-A | HET | ||
| SBS | Chr2 | 26330205 | G-A | HET | ||
| SBS | Chr2 | 28566880 | T-A | HET | ||
| SBS | Chr2 | 28566881 | T-A | HET | ||
| SBS | Chr2 | 28566911 | C-G | HET | ||
| SBS | Chr2 | 5366959 | T-G | Homo | ||
| SBS | Chr3 | 12491478 | T-A | HET | ||
| SBS | Chr3 | 13441053 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g23220 |
| SBS | Chr3 | 16365730 | G-T | HET | ||
| SBS | Chr3 | 17192676 | A-G | HET | ||
| SBS | Chr3 | 21198128 | A-T | Homo | ||
| SBS | Chr3 | 24246342 | C-T | HET | ||
| SBS | Chr3 | 2640353 | T-C | HET | ||
| SBS | Chr3 | 29471887 | C-A | HET | ||
| SBS | Chr4 | 12484914 | G-A | HET | ||
| SBS | Chr4 | 12484915 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g22050 |
| SBS | Chr4 | 13385039 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g23410 |
| SBS | Chr5 | 10432664 | T-A | HET | ||
| SBS | Chr6 | 13863408 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g23720 |
| SBS | Chr6 | 30414172 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g50220 |
| SBS | Chr6 | 31208227 | C-T | HET | ||
| SBS | Chr6 | 774167 | G-A | HET | ||
| SBS | Chr7 | 10310218 | G-A | HET | ||
| SBS | Chr7 | 25870686 | A-C | Homo | ||
| SBS | Chr7 | 2628562 | G-A | HET | ||
| SBS | Chr8 | 15784328 | C-A | HET | ||
| SBS | Chr8 | 16575016 | C-T | HET | ||
| SBS | Chr8 | 27220587 | C-A | HET | ||
| SBS | Chr8 | 4842731 | G-A | HET | ||
| SBS | Chr9 | 11875681 | A-C | HET | ||
| SBS | Chr9 | 1629509 | C-T | Homo | ||
| SBS | Chr9 | 20764755 | C-T | HET | ||
| SBS | Chr9 | 21666633 | T-A | HET |
Deletions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 5315929 | 5315941 | 12 | LOC_Os12g10030 |
| Deletion | Chr3 | 9187117 | 9187129 | 12 | |
| Deletion | Chr8 | 14788830 | 14788838 | 8 | LOC_Os08g24480 |
| Deletion | Chr7 | 16203001 | 16449000 | 245999 | 40 |
| Deletion | Chr12 | 17910978 | 17910991 | 13 | |
| Deletion | Chr9 | 18646653 | 18646656 | 3 | |
| Deletion | Chr6 | 26149694 | 26149697 | 3 | |
| Deletion | Chr4 | 27023737 | 27023739 | 2 | |
| Deletion | Chr5 | 27458078 | 27458080 | 2 | LOC_Os05g47880 |
| Deletion | Chr4 | 30898073 | 30898077 | 4 | LOC_Os04g52020 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 15188015 | 15188017 | 3 | |
| Insertion | Chr2 | 34727403 | 34727403 | 1 | LOC_Os02g56640 |
| Insertion | Chr2 | 9009052 | 9009150 | 99 | |
| Insertion | Chr5 | 25602269 | 25602269 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 1052106 | 1052450 | |
| Inversion | Chr4 | 11560408 | 11750970 | LOC_Os04g20940 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 7841089 | Chr1 | 219871 | LOC_Os01g01440 |
| Translocation | Chr11 | 13573527 | Chr4 | 7015308 | LOC_Os04g12669 |
| Translocation | Chr10 | 16098010 | Chr7 | 4798226 | LOC_Os10g30870 |
| Translocation | Chr11 | 28683193 | Chr7 | 1481182 |
