Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4344-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4344-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 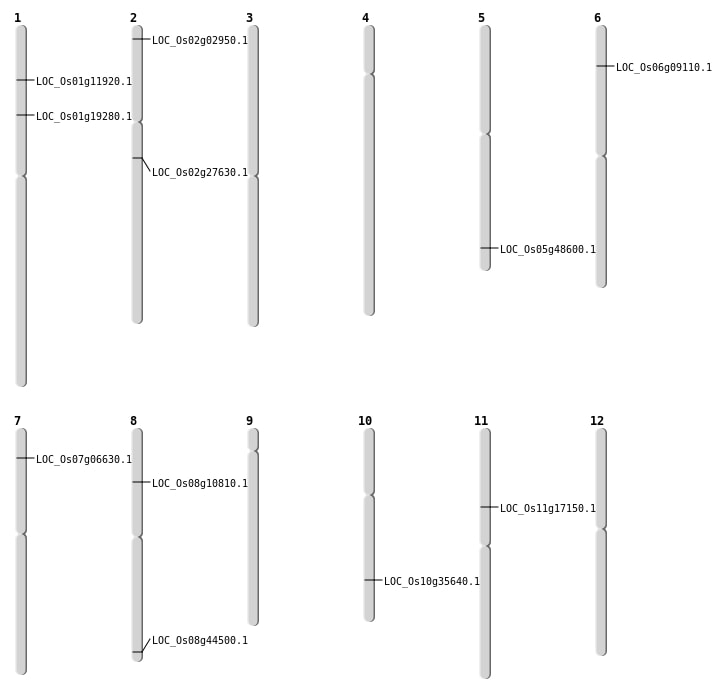
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10900049 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g19280 |
| SBS | Chr1 | 21968749 | G-A | HET | ||
| SBS | Chr1 | 3393632 | G-A | Homo | ||
| SBS | Chr1 | 8144979 | C-A | HET | ||
| SBS | Chr10 | 22851814 | T-A | HET | ||
| SBS | Chr10 | 4207581 | A-T | Homo | ||
| SBS | Chr11 | 12821916 | T-C | HET | ||
| SBS | Chr11 | 16930853 | T-A | HET | ||
| SBS | Chr11 | 24927066 | G-T | HET | ||
| SBS | Chr11 | 8980135 | G-A | HET | ||
| SBS | Chr11 | 9523418 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g17150 |
| SBS | Chr12 | 15871413 | C-T | HET | ||
| SBS | Chr12 | 17515879 | C-T | HET | ||
| SBS | Chr12 | 25750378 | G-A | HET | ||
| SBS | Chr2 | 1155332 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g02950 |
| SBS | Chr2 | 1320260 | A-T | Homo | ||
| SBS | Chr2 | 14055269 | C-T | HET | ||
| SBS | Chr2 | 23021748 | G-A | Homo | ||
| SBS | Chr2 | 24912223 | G-A | Homo | ||
| SBS | Chr2 | 926859 | C-T | Homo | ||
| SBS | Chr3 | 16761902 | G-A | Homo | ||
| SBS | Chr3 | 487107 | C-T | Homo | ||
| SBS | Chr3 | 702905 | A-G | HET | ||
| SBS | Chr4 | 28083105 | T-G | HET | ||
| SBS | Chr4 | 28083106 | A-T | HET | ||
| SBS | Chr4 | 28083107 | G-A | HET | ||
| SBS | Chr4 | 32708702 | G-A | HET | ||
| SBS | Chr4 | 33054571 | G-T | HET | ||
| SBS | Chr5 | 16353638 | G-A | HET | ||
| SBS | Chr5 | 19020844 | G-C | Homo | ||
| SBS | Chr5 | 19716798 | T-A | HET | ||
| SBS | Chr5 | 2147210 | T-C | HET | ||
| SBS | Chr5 | 22891557 | A-T | HET | ||
| SBS | Chr6 | 22221542 | G-A | HET | ||
| SBS | Chr6 | 22221543 | G-A | HET | ||
| SBS | Chr6 | 22432299 | G-A | HET | ||
| SBS | Chr6 | 30436203 | G-A | Homo | ||
| SBS | Chr7 | 20783800 | G-A | Homo | ||
| SBS | Chr8 | 13777220 | G-A | HET | ||
| SBS | Chr8 | 24535622 | C-T | Homo | ||
| SBS | Chr8 | 27987852 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g44500 |
| SBS | Chr8 | 8886459 | T-A | HET | ||
| SBS | Chr9 | 12822699 | T-A | HET | ||
| SBS | Chr9 | 1508108 | T-A | HET | ||
| SBS | Chr9 | 16537384 | C-T | HET |
Deletions: 9
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 1698393 | 1698394 | 1 | |
| Deletion | Chr11 | 3919201 | 3919204 | 3 | |
| Deletion | Chr6 | 4583918 | 4583923 | 5 | LOC_Os06g09110 |
| Deletion | Chr1 | 10323318 | 10323325 | 7 | |
| Deletion | Chr2 | 10328581 | 10328598 | 17 | |
| Deletion | Chr2 | 16375468 | 16375474 | 6 | LOC_Os02g27630 |
| Deletion | Chr10 | 16396595 | 16396601 | 6 | |
| Deletion | Chr10 | 19058076 | 19058077 | 1 | LOC_Os10g35640 |
| Deletion | Chr11 | 20247544 | 20247545 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 18079442 | 18079444 | 3 | |
| Insertion | Chr3 | 13386871 | 13386871 | 1 | |
| Insertion | Chr5 | 13988934 | 13989040 | 107 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 6368093 | 6940218 | LOC_Os08g10810 |
| Inversion | Chr8 | 6733846 | 6734212 | |
| Inversion | Chr1 | 10397137 | 11083624 | |
| Inversion | Chr5 | 27862989 | 28143582 | LOC_Os05g48600 |
| Inversion | Chr5 | 27862994 | 28143585 | LOC_Os05g48600 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 3231080 | Chr1 | 6479370 | 2 |
| Translocation | Chr4 | 4118420 | Chr3 | 21465998 | |
| Translocation | Chr12 | 15876581 | Chr2 | 16818007 |
