Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4354-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4354-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 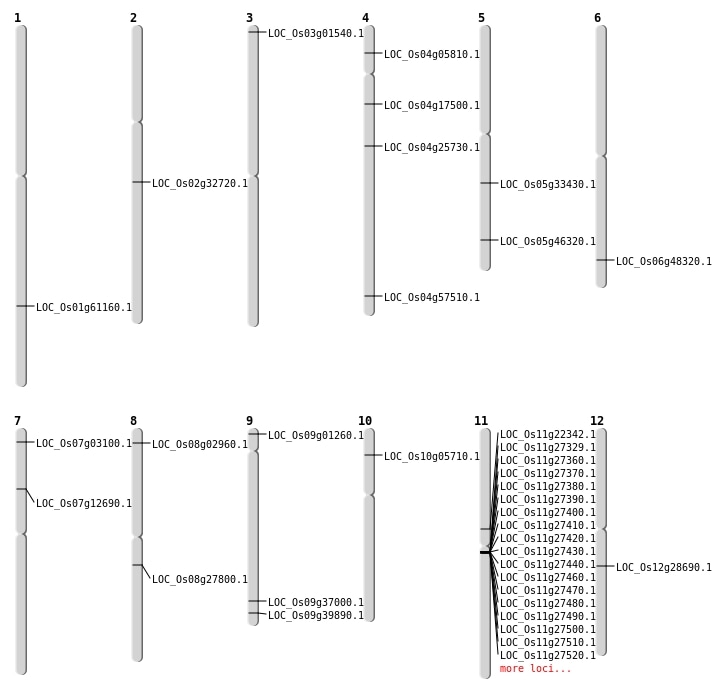
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11841335 | C-T | HET | INTRON | |
| SBS | Chr1 | 30252166 | A-T | HET | INTRON | |
| SBS | Chr1 | 38396289 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 2879960 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g05710 |
| SBS | Chr11 | 10421449 | A-C | HOMO | INTRON | |
| SBS | Chr11 | 12807232 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g22342 |
| SBS | Chr11 | 17209158 | A-G | HET | UTR_5_PRIME | |
| SBS | Chr12 | 18824882 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 1885608 | A-G | HET | INTRON | |
| SBS | Chr12 | 5286339 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 19433833 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g32720 |
| SBS | Chr2 | 27172839 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 13395197 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 6617486 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 12520241 | G-T | HET | INTRON | |
| SBS | Chr4 | 14913863 | C-T | HET | STOP_GAINED | LOC_Os04g25730 |
| SBS | Chr4 | 14935725 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 1704244 | T-A | HET | INTRON | |
| SBS | Chr4 | 2985028 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g05810 |
| SBS | Chr4 | 34211889 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g57510 |
| SBS | Chr5 | 10461996 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 10461997 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 26847138 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g46320 |
| SBS | Chr6 | 10677023 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 13536942 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 15073735 | G-A | HET | INTRON | |
| SBS | Chr6 | 15397684 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 28786824 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 1181003 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g03100 |
| SBS | Chr7 | 4363727 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 12972727 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 16285428 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 16343040 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 21339566 | A-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g37000 |
| SBS | Chr9 | 4931640 | C-T | HET | INTRON | |
| SBS | Chr9 | 5758995 | C-T | HET | INTERGENIC |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 345146 | 345152 | 6 | LOC_Os03g01540 |
| Deletion | Chr8 | 1288043 | 1288059 | 16 | LOC_Os08g02960 |
| Deletion | Chr12 | 4233440 | 4233441 | 1 | |
| Deletion | Chr6 | 5564113 | 5564153 | 40 | |
| Deletion | Chr9 | 6380058 | 6380059 | 1 | |
| Deletion | Chr8 | 7267712 | 7267713 | 1 | |
| Deletion | Chr8 | 8545561 | 8545562 | 1 | |
| Deletion | Chr10 | 9111421 | 9111422 | 1 | |
| Deletion | Chr4 | 9571723 | 9571724 | 1 | LOC_Os04g17500 |
| Deletion | Chr6 | 11098983 | 11098985 | 2 | |
| Deletion | Chr11 | 12111336 | 12111395 | 59 | |
| Deletion | Chr11 | 14471844 | 14471857 | 13 | |
| Deletion | Chr5 | 15442509 | 15442511 | 2 | |
| Deletion | Chr10 | 15519744 | 15519807 | 63 | |
| Deletion | Chr11 | 15715001 | 15770000 | 55000 | 6 |
| Deletion | Chr11 | 15775001 | 15881000 | 106000 | 16 |
| Deletion | Chr3 | 15835442 | 15835443 | 1 | |
| Deletion | Chr8 | 16933189 | 16933192 | 3 | LOC_Os08g27800 |
| Deletion | Chr12 | 16952123 | 16952182 | 59 | LOC_Os12g28690 |
| Deletion | Chr9 | 17189192 | 17189261 | 69 | |
| Deletion | Chr5 | 19646748 | 19646763 | 15 | LOC_Os05g33430 |
| Deletion | Chr2 | 20418781 | 20418783 | 2 | |
| Deletion | Chr9 | 22870321 | 22870345 | 24 | LOC_Os09g39890 |
| Deletion | Chr11 | 27289179 | 27289193 | 14 | |
| Deletion | Chr4 | 29249506 | 29249507 | 1 | |
| Deletion | Chr2 | 33133286 | 33133287 | 1 | |
| Deletion | Chr1 | 35404500 | 35404501 | 1 | LOC_Os01g61160 |
| Deletion | Chr1 | 42468540 | 42468610 | 70 |
Insertions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 23693715 | 23693717 | 3 | |
| Insertion | Chr12 | 24073238 | 24073239 | 2 | |
| Insertion | Chr12 | 50432 | 50432 | 1 | |
| Insertion | Chr3 | 10897436 | 10897437 | 2 | |
| Insertion | Chr3 | 17060881 | 17060881 | 1 | |
| Insertion | Chr4 | 10422845 | 10422845 | 1 | |
| Insertion | Chr6 | 25488478 | 25488478 | 1 | |
| Insertion | Chr7 | 7252105 | 7252105 | 1 | LOC_Os07g12690 |
| Insertion | Chr7 | 9704209 | 9704209 | 1 | |
| Insertion | Chr8 | 11039012 | 11039023 | 12 | |
| Insertion | Chr8 | 1332304 | 1332304 | 1 | |
| Insertion | Chr8 | 13388316 | 13388318 | 3 | |
| Insertion | Chr9 | 18082131 | 18082152 | 22 | |
| Insertion | Chr9 | 21898030 | 21898030 | 1 | |
| Insertion | Chr9 | 22006664 | 22006667 | 4 | |
| Insertion | Chr9 | 223262 | 223262 | 1 | LOC_Os09g01260 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 3080006 | 3080391 | |
| Inversion | Chr6 | 3080008 | 3080397 | |
| Inversion | Chr10 | 8780856 | 8780938 | |
| Inversion | Chr5 | 22517650 | 22517840 | |
| Inversion | Chr6 | 29217510 | 29217722 | LOC_Os06g48320 |
No Translocation
