Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4377-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4377-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 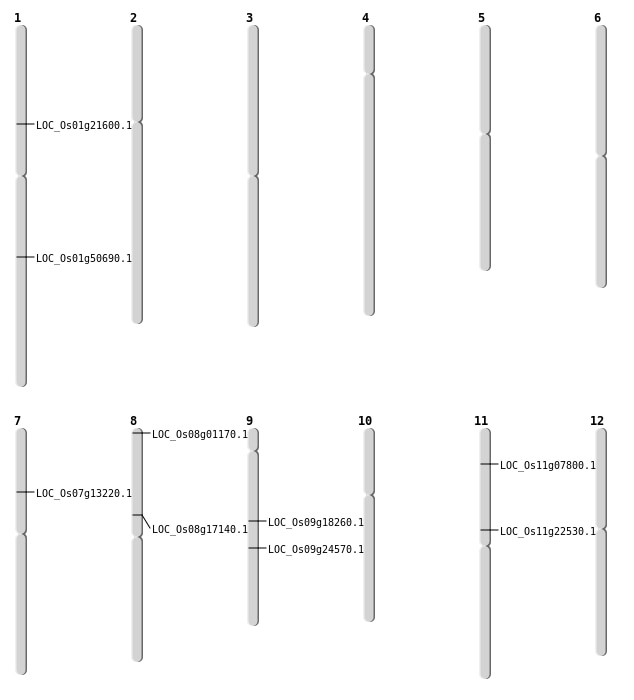
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10828062 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 21778308 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 22162313 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 13047121 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 1812725 | G-A | HET | INTRON | |
| SBS | Chr11 | 18773314 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 1986937 | C-A | HET | INTRON | |
| SBS | Chr11 | 7969526 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 14725240 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 18201695 | G-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 22961258 | G-A | HET | INTRON | |
| SBS | Chr2 | 23508454 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 23785751 | G-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 24938267 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 26542572 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 28556842 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 31601017 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 3731022 | G-C | HET | UTR_3_PRIME | |
| SBS | Chr4 | 10849948 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 17686708 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 23599367 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 29285836 | T-G | HOMO | INTRON | |
| SBS | Chr4 | 5535478 | T-C | HOMO | INTRON | |
| SBS | Chr5 | 23498493 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 26856685 | T-G | HET | UTR_3_PRIME | |
| SBS | Chr5 | 27077439 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 11839194 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 5327073 | G-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 5327074 | A-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 6264554 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 6264556 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 6036808 | T-A | HET | INTRON | |
| SBS | Chr7 | 7575664 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g13220 |
| SBS | Chr8 | 109795 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g01170 |
| SBS | Chr8 | 4290468 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr8 | 4781332 | A-G | HET | INTRON | |
| SBS | Chr8 | 5792986 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 11212013 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g18260 |
| SBS | Chr9 | 14631541 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g24570 |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 771017 | 771018 | 1 | |
| Deletion | Chr11 | 790634 | 790641 | 7 | |
| Deletion | Chr2 | 1934754 | 1934755 | 1 | |
| Deletion | Chr11 | 3435471 | 3435472 | 1 | |
| Deletion | Chr12 | 3832468 | 3832470 | 2 | |
| Deletion | Chr11 | 3983618 | 3983619 | 1 | LOC_Os11g07800 |
| Deletion | Chr8 | 4293650 | 4293651 | 1 | |
| Deletion | Chr3 | 4336513 | 4336514 | 1 | |
| Deletion | Chr8 | 4754318 | 4754319 | 1 | |
| Deletion | Chr12 | 5344461 | 5344462 | 1 | |
| Deletion | Chr7 | 7013246 | 7013256 | 10 | |
| Deletion | Chr1 | 9295718 | 9295719 | 1 | |
| Deletion | Chr11 | 10079776 | 10079781 | 5 | |
| Deletion | Chr12 | 10729882 | 10729913 | 31 | |
| Deletion | Chr7 | 11613268 | 11613269 | 1 | |
| Deletion | Chr11 | 12942365 | 12942367 | 2 | LOC_Os11g22530 |
| Deletion | Chr8 | 14440092 | 14440094 | 2 | |
| Deletion | Chr2 | 15569108 | 15569109 | 1 | |
| Deletion | Chr12 | 16890531 | 16890546 | 15 | |
| Deletion | Chr11 | 18237388 | 18237389 | 1 | |
| Deletion | Chr11 | 20051423 | 20051429 | 6 | |
| Deletion | Chr4 | 20933866 | 20933901 | 35 | |
| Deletion | Chr6 | 23246944 | 23246964 | 20 | |
| Deletion | Chr8 | 27348688 | 27348689 | 1 | |
| Deletion | Chr7 | 27373654 | 27373655 | 1 | |
| Deletion | Chr6 | 29012517 | 29012611 | 94 | |
| Deletion | Chr6 | 30475783 | 30475784 | 1 |
Insertions: 10
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 8145971 | 8527595 | |
| Inversion | Chr12 | 9863157 | 10075193 | |
| Inversion | Chr8 | 10494066 | 11482624 | LOC_Os08g17140 |
| Inversion | Chr8 | 10494067 | 11482625 | LOC_Os08g17140 |
| Inversion | Chr4 | 17788913 | 17789068 | |
| Inversion | Chr1 | 29113165 | 29113297 | LOC_Os01g50690 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 2253482 | Chr4 | 1081454 | |
| Translocation | Chr12 | 12101166 | Chr1 | 17417127 | LOC_Os01g21600 |
| Translocation | Chr10 | 15534760 | Chr1 | 23827307 | |
| Translocation | Chr6 | 22360143 | Chr1 | 26412731 | |
| Translocation | Chr12 | 22853357 | Chr1 | 15881136 |
