Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4378-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4378-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 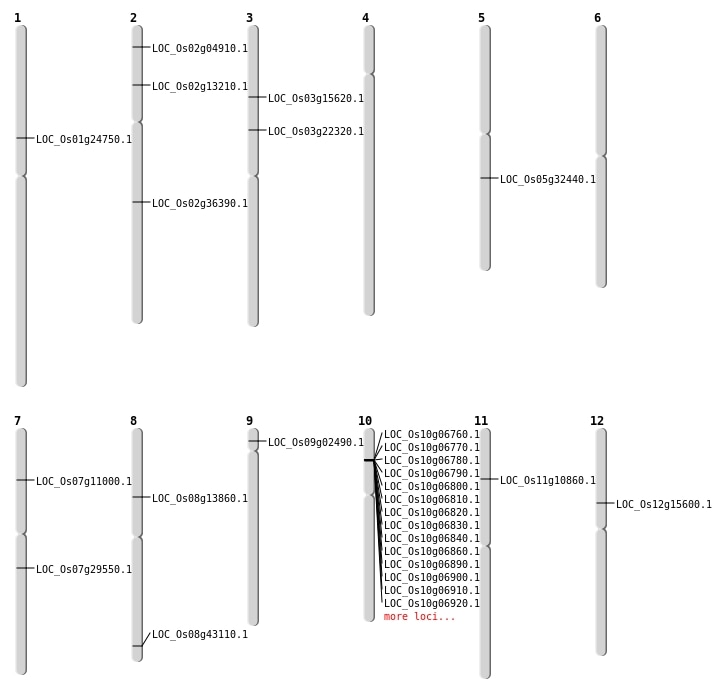
|
| Variant Information |
|---|
Single base substitutions: 24
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13924323 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g24750 |
| SBS | Chr1 | 35662933 | G-A | HET | INTRON | |
| SBS | Chr1 | 35662934 | A-T | HET | INTRON | |
| SBS | Chr1 | 9581358 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 11630618 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g22460 |
| SBS | Chr10 | 20064541 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr10 | 6519796 | G-A | HET | INTRON | |
| SBS | Chr12 | 8916227 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g15600 |
| SBS | Chr2 | 7779246 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 12789767 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g22320 |
| SBS | Chr3 | 16194474 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 18260277 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 13532525 | A-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 9781703 | C-G | HOMO | INTRON | |
| SBS | Chr5 | 12079742 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 12079743 | T-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 18942714 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g32440 |
| SBS | Chr6 | 24271114 | C-T | HET | INTRON | |
| SBS | Chr7 | 17349628 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g29550 |
| SBS | Chr8 | 24505321 | A-G | HOMO | INTRON | |
| SBS | Chr8 | 4230500 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 8278568 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g13860 |
| SBS | Chr9 | 1032118 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g02490 |
| SBS | Chr9 | 18671126 | G-C | HET | UTR_3_PRIME |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 707764 | 707765 | 1 | |
| Deletion | Chr12 | 730875 | 730887 | 12 | |
| Deletion | Chr2 | 2273031 | 2273034 | 3 | LOC_Os02g04910 |
| Deletion | Chr1 | 2718788 | 2718789 | 1 | |
| Deletion | Chr10 | 3511001 | 4065000 | 554000 | 73 |
| Deletion | Chr12 | 3538441 | 3538442 | 1 | |
| Deletion | Chr3 | 3545625 | 3545626 | 1 | |
| Deletion | Chr11 | 5982724 | 5982727 | 3 | LOC_Os11g10860 |
| Deletion | Chr5 | 7281389 | 7281390 | 1 | |
| Deletion | Chr8 | 7861792 | 7861809 | 17 | |
| Deletion | Chr3 | 8618704 | 8618705 | 1 | LOC_Os03g15620 |
| Deletion | Chr5 | 8765184 | 8765188 | 4 | |
| Deletion | Chr4 | 9452182 | 9452184 | 2 | |
| Deletion | Chr4 | 11522832 | 11522833 | 1 | |
| Deletion | Chr6 | 15431496 | 15431497 | 1 | |
| Deletion | Chr6 | 16125664 | 16125674 | 10 | |
| Deletion | Chr11 | 17987206 | 17987207 | 1 | |
| Deletion | Chr11 | 20220875 | 20220876 | 1 | |
| Deletion | Chr9 | 21429585 | 21429586 | 1 | |
| Deletion | Chr3 | 21571146 | 21571147 | 1 | |
| Deletion | Chr10 | 22218504 | 22218505 | 1 | |
| Deletion | Chr11 | 22647287 | 22647292 | 5 | |
| Deletion | Chr1 | 22704711 | 22704712 | 1 | |
| Deletion | Chr1 | 23918359 | 23918366 | 7 | |
| Deletion | Chr1 | 25296061 | 25296152 | 91 | |
| Deletion | Chr12 | 26436606 | 26436607 | 1 | |
| Deletion | Chr4 | 27007832 | 27007834 | 2 | |
| Deletion | Chr12 | 27138387 | 27138388 | 1 | |
| Deletion | Chr6 | 27831435 | 27831437 | 2 | |
| Deletion | Chr2 | 29860832 | 29860835 | 3 | |
| Deletion | Chr3 | 36068408 | 36068409 | 1 | |
| Deletion | Chr1 | 42765131 | 42765149 | 18 |
Insertions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 10535949 | 10535949 | 1 | |
| Insertion | Chr1 | 3021737 | 3021746 | 10 | |
| Insertion | Chr10 | 16597103 | 16597103 | 1 | |
| Insertion | Chr11 | 8869793 | 8869793 | 1 | |
| Insertion | Chr12 | 11913766 | 11913767 | 2 | |
| Insertion | Chr12 | 17542069 | 17542069 | 1 | |
| Insertion | Chr12 | 3105418 | 3105418 | 1 | |
| Insertion | Chr2 | 16273513 | 16273554 | 42 | |
| Insertion | Chr2 | 19722828 | 19722834 | 7 | |
| Insertion | Chr2 | 29417037 | 29417037 | 1 | |
| Insertion | Chr2 | 32724924 | 32724925 | 2 | |
| Insertion | Chr2 | 7040501 | 7040501 | 1 | LOC_Os02g13210 |
| Insertion | Chr2 | 7523640 | 7523640 | 1 | |
| Insertion | Chr3 | 28140175 | 28140175 | 1 | |
| Insertion | Chr3 | 35720619 | 35720619 | 1 | |
| Insertion | Chr3 | 36309216 | 36309217 | 2 | |
| Insertion | Chr3 | 9059126 | 9059128 | 3 | |
| Insertion | Chr4 | 33500758 | 33500759 | 2 | |
| Insertion | Chr5 | 13350245 | 13350245 | 1 | |
| Insertion | Chr6 | 9006320 | 9006320 | 1 | |
| Insertion | Chr7 | 6931447 | 6931448 | 2 | |
| Insertion | Chr8 | 27255845 | 27255845 | 1 | LOC_Os08g43110 |
| Insertion | Chr8 | 28344486 | 28344486 | 1 | |
| Insertion | Chr8 | 8017385 | 8017394 | 10 | |
| Insertion | Chr9 | 4919476 | 4919476 | 1 | |
| Insertion | Chr9 | 8441972 | 8441975 | 4 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 6039940 | 6040015 | LOC_Os07g11000 |
| Inversion | Chr2 | 21983334 | 21983624 | LOC_Os02g36390 |
No Translocation
