Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN438-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN438-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 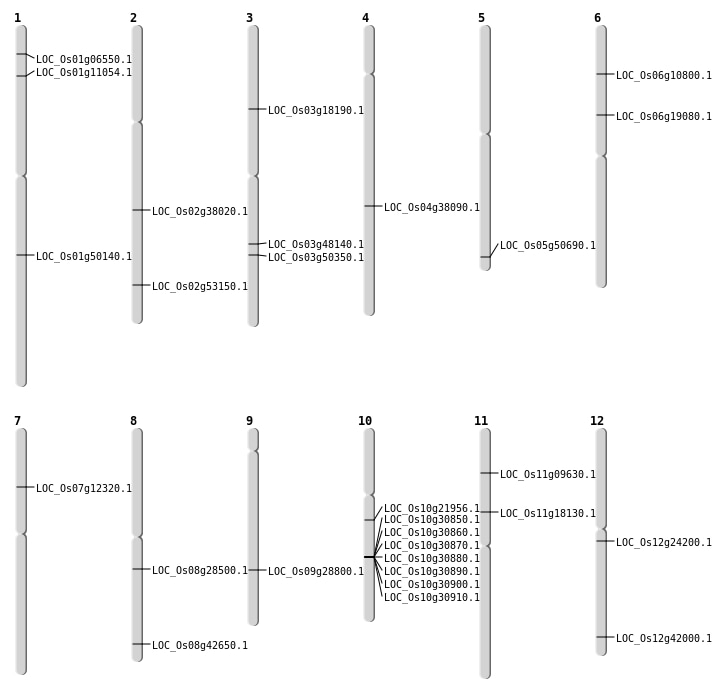
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 2352795 | C-A | HET | INTRON | |
| SBS | Chr1 | 28351078 | C-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 5907055 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g11054 |
| SBS | Chr10 | 10091489 | A-T | HET | INTRON | |
| SBS | Chr10 | 12400216 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 12738525 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 16814499 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 2285239 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 9245966 | G-A | HET | INTRON | |
| SBS | Chr12 | 21660384 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 19605954 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 32635123 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 19097787 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 25502525 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 27348174 | C-T | HET | INTRON | |
| SBS | Chr3 | 5776600 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 5776605 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 5776613 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 11595927 | C-G | HET | INTRON | |
| SBS | Chr4 | 21415473 | A-T | HET | INTRON | |
| SBS | Chr4 | 22673090 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g38090 |
| SBS | Chr4 | 27857503 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 24392801 | C-T | HOMO | INTRON | |
| SBS | Chr6 | 10628973 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 13082508 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 26964117 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 26964118 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 22970618 | A-G | HOMO | INTRON | |
| SBS | Chr7 | 24599700 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 10166822 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 17399705 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g28500 |
| SBS | Chr8 | 26960215 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g42650 |
| SBS | Chr8 | 8253819 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 17495224 | G-C | HET | STOP_GAINED | LOC_Os09g28800 |
| SBS | Chr9 | 22837421 | C-T | HET | INTRON |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 1658398 | 1658408 | 10 | |
| Deletion | Chr5 | 1712671 | 1712677 | 6 | |
| Deletion | Chr5 | 1824596 | 1824597 | 1 | |
| Deletion | Chr1 | 3085904 | 3085921 | 17 | LOC_Os01g06550 |
| Deletion | Chr12 | 3242061 | 3242062 | 1 | |
| Deletion | Chr11 | 5143184 | 5148538 | 5354 | LOC_Os11g09630 |
| Deletion | Chr6 | 5637116 | 5637117 | 1 | LOC_Os06g10800 |
| Deletion | Chr5 | 6475245 | 6475246 | 1 | |
| Deletion | Chr7 | 6863540 | 6863545 | 5 | |
| Deletion | Chr7 | 6951746 | 6951753 | 7 | LOC_Os07g12320 |
| Deletion | Chr1 | 8173399 | 8173400 | 1 | |
| Deletion | Chr1 | 8414990 | 8414993 | 3 | |
| Deletion | Chr3 | 10195624 | 10195625 | 1 | LOC_Os03g18190 |
| Deletion | Chr8 | 13122459 | 13122468 | 9 | |
| Deletion | Chr12 | 13794138 | 13794139 | 1 | LOC_Os12g24200 |
| Deletion | Chr3 | 13801216 | 13801218 | 2 | |
| Deletion | Chr10 | 15558452 | 15558455 | 3 | |
| Deletion | Chr5 | 15725764 | 15725769 | 5 | |
| Deletion | Chr10 | 15912065 | 15912066 | 1 | |
| Deletion | Chr10 | 16071001 | 16122000 | 51000 | 7 |
| Deletion | Chr7 | 17057367 | 17057370 | 3 | |
| Deletion | Chr9 | 21617891 | 21617897 | 6 | |
| Deletion | Chr2 | 22469891 | 22469921 | 30 | |
| Deletion | Chr2 | 22974326 | 22974335 | 9 | LOC_Os02g38020 |
| Deletion | Chr3 | 24168123 | 24168496 | 373 | |
| Deletion | Chr1 | 25186738 | 25186744 | 6 | |
| Deletion | Chr12 | 26044774 | 26044779 | 5 | LOC_Os12g42000 |
| Deletion | Chr3 | 26054591 | 26054605 | 14 | |
| Deletion | Chr8 | 26259092 | 26259094 | 2 | |
| Deletion | Chr3 | 27385193 | 27385299 | 106 | LOC_Os03g48140 |
| Deletion | Chr3 | 28731224 | 28731227 | 3 | LOC_Os03g50350 |
| Deletion | Chr1 | 28808603 | 28808624 | 21 | LOC_Os01g50140 |
| Deletion | Chr5 | 29061533 | 29061537 | 4 | LOC_Os05g50690 |
| Deletion | Chr2 | 32535804 | 32535820 | 16 | LOC_Os02g53150 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 17086521 | 17086521 | 1 | |
| Insertion | Chr3 | 21038827 | 21038827 | 1 | |
| Insertion | Chr9 | 20166224 | 20166225 | 2 | |
| Insertion | Chr9 | 9495466 | 9495467 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 9912609 | 10204270 | LOC_Os11g18130 |
| Inversion | Chr11 | 9912632 | 10204298 | LOC_Os11g18130 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 9912596 | Chr3 | 24168493 | |
| Translocation | Chr11 | 10204263 | Chr3 | 24168135 | LOC_Os11g18130 |
| Translocation | Chr10 | 11338810 | Chr6 | 10850161 | 2 |
