Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4380-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4380-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 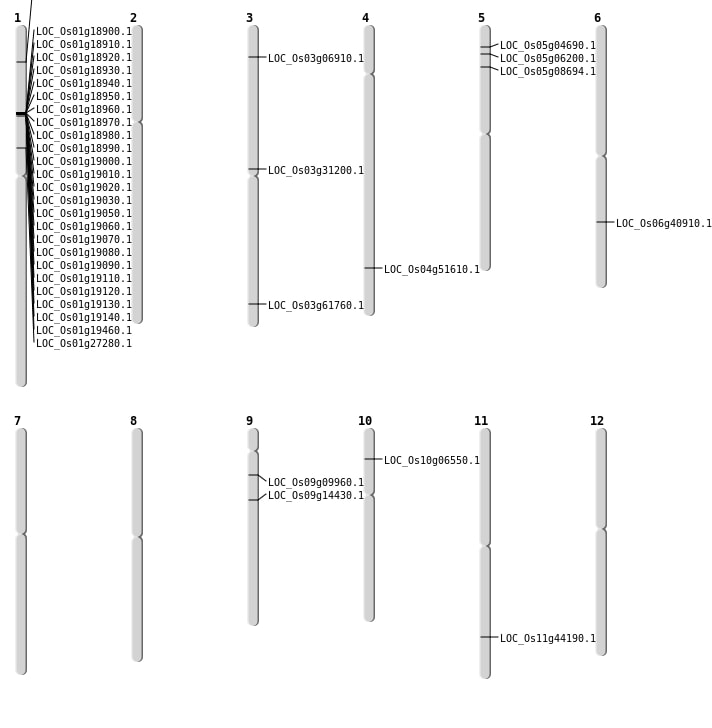
|
| Variant Information |
|---|
Single base substitutions: 28
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11019646 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g19460 |
| SBS | Chr1 | 24298189 | C-T | HET | INTRON | |
| SBS | Chr1 | 36133473 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 20096232 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 7218197 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 8079720 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 14724329 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 753757 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 13369808 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 290500 | T-G | HET | INTRON | |
| SBS | Chr2 | 8839446 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 24227943 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 3471529 | A-C | HOMO | UTR_3_PRIME | |
| SBS | Chr4 | 30574718 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g51610 |
| SBS | Chr4 | 33018955 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 23777631 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 3120294 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g06200 |
| SBS | Chr5 | 4764593 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g08694 |
| SBS | Chr5 | 4847804 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 21306485 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 23766802 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 27255344 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 21133877 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 4378164 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 13325319 | T-C | HET | INTRON | |
| SBS | Chr9 | 5421620 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g09960 |
| SBS | Chr9 | 5427489 | G-A | HOMO | INTRON | |
| SBS | Chr9 | 8524676 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g14430 |
Deletions: 37
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 939350 | 939355 | 5 | |
| Deletion | Chr5 | 2217664 | 2217676 | 12 | LOC_Os05g04690 |
| Deletion | Chr3 | 2710226 | 2710229 | 3 | |
| Deletion | Chr8 | 2970804 | 2970809 | 5 | |
| Deletion | Chr10 | 3368119 | 3368126 | 7 | LOC_Os10g06550 |
| Deletion | Chr3 | 3503229 | 3503230 | 1 | LOC_Os03g06910 |
| Deletion | Chr10 | 4430060 | 4430061 | 1 | |
| Deletion | Chr1 | 4492102 | 4492103 | 1 | |
| Deletion | Chr12 | 5447822 | 5447823 | 1 | |
| Deletion | Chr9 | 5863401 | 5863403 | 2 | |
| Deletion | Chr10 | 7793656 | 7793657 | 1 | |
| Deletion | Chr9 | 9361118 | 9361198 | 80 | |
| Deletion | Chr1 | 10683001 | 10725000 | 42000 | 9 |
| Deletion | Chr1 | 10728001 | 10789000 | 61000 | 10 |
| Deletion | Chr1 | 10794001 | 10819000 | 25000 | 4 |
| Deletion | Chr7 | 13604503 | 13604504 | 1 | |
| Deletion | Chr1 | 15223212 | 15223213 | 1 | LOC_Os01g27280 |
| Deletion | Chr12 | 16044066 | 16044067 | 1 | |
| Deletion | Chr3 | 17663190 | 17663191 | 1 | |
| Deletion | Chr3 | 17766487 | 17766488 | 1 | LOC_Os03g31200 |
| Deletion | Chr7 | 18316285 | 18316286 | 1 | |
| Deletion | Chr5 | 18649456 | 18649457 | 1 | |
| Deletion | Chr11 | 20062534 | 20062540 | 6 | |
| Deletion | Chr12 | 20224514 | 20224515 | 1 | |
| Deletion | Chr6 | 21053432 | 21053434 | 2 | |
| Deletion | Chr4 | 21195198 | 21195199 | 1 | |
| Deletion | Chr11 | 21224505 | 21224507 | 2 | |
| Deletion | Chr1 | 22378139 | 22378140 | 1 | |
| Deletion | Chr8 | 23421995 | 23421996 | 1 | |
| Deletion | Chr11 | 26689768 | 26689769 | 1 | LOC_Os11g44190 |
| Deletion | Chr7 | 26827508 | 26827509 | 1 | |
| Deletion | Chr8 | 27913160 | 27913161 | 1 | |
| Deletion | Chr1 | 31773962 | 31773963 | 1 | |
| Deletion | Chr3 | 32638902 | 32638908 | 6 | |
| Deletion | Chr2 | 34714570 | 34714571 | 1 | |
| Deletion | Chr4 | 35063028 | 35063029 | 1 | |
| Deletion | Chr3 | 36327826 | 36327827 | 1 |
Insertions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 4211128 | 4211130 | 3 | LOC_Os01g08510 |
| Insertion | Chr11 | 23250833 | 23250840 | 8 | |
| Insertion | Chr12 | 11626674 | 11626674 | 1 | |
| Insertion | Chr2 | 19065392 | 19065392 | 1 | |
| Insertion | Chr2 | 32948121 | 32948121 | 1 | |
| Insertion | Chr3 | 11659449 | 11659449 | 1 | |
| Insertion | Chr3 | 35012181 | 35012181 | 1 | LOC_Os03g61760 |
| Insertion | Chr4 | 33279089 | 33279089 | 1 | |
| Insertion | Chr6 | 30919679 | 30919679 | 1 | |
| Insertion | Chr7 | 16854379 | 16854386 | 8 | |
| Insertion | Chr7 | 24850982 | 24851044 | 63 | |
| Insertion | Chr8 | 11235069 | 11235069 | 1 | |
| Insertion | Chr9 | 7872350 | 7872350 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 5979718 | 5979976 | |
| Inversion | Chr6 | 28833620 | 28833692 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 6904424 | Chr1 | 11767719 | |
| Translocation | Chr5 | 6904433 | Chr1 | 11768118 | |
| Translocation | Chr7 | 24388510 | Chr6 | 16837848 | LOC_Os06g40910 |
