Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4381-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4381-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 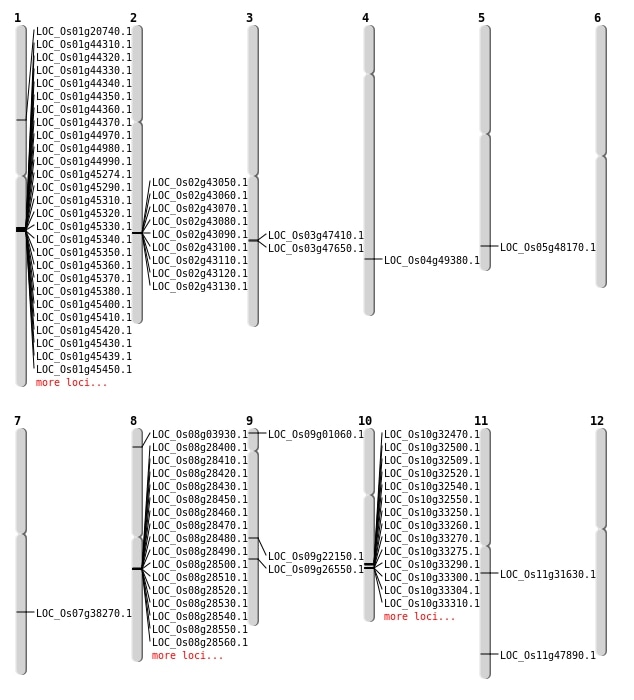
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 5986547 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 17603407 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 20639442 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 22802378 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 11179822 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 11743295 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 18497756 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 18497757 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g31630 |
| SBS | Chr11 | 219416 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 28888665 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g47890 |
| SBS | Chr12 | 10815674 | C-T | HET | INTRON | |
| SBS | Chr12 | 12978280 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 14225872 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 20408827 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 27249519 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 8199267 | C-T | HET | INTRON | |
| SBS | Chr2 | 17333187 | A-G | HET | INTRON | |
| SBS | Chr2 | 20255043 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 26392004 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 20119275 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 26802161 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g47410 |
| SBS | Chr3 | 26989518 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g47650 |
| SBS | Chr4 | 15853360 | G-C | HOMO | UTR_5_PRIME | |
| SBS | Chr4 | 4090466 | T-C | HET | INTRON | |
| SBS | Chr5 | 10535690 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 5592478 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 9642298 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 13321032 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 18099203 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 21112784 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 5243809 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 15139695 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 18226425 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 25588774 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 29385019 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr7 | 7494048 | G-T | HET | INTRON | |
| SBS | Chr8 | 13149179 | G-A | HET | INTRON | |
| SBS | Chr9 | 13408479 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g22150 |
| SBS | Chr9 | 19131612 | C-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 8362256 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 89146 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g01060 |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 430620 | 430622 | 2 | |
| Deletion | Chr8 | 1867956 | 1867958 | 2 | LOC_Os08g03930 |
| Deletion | Chr6 | 2826123 | 2826135 | 12 | |
| Deletion | Chr3 | 9118174 | 9118214 | 40 | |
| Deletion | Chr7 | 9239980 | 9239981 | 1 | |
| Deletion | Chr1 | 10186488 | 10186490 | 2 | |
| Deletion | Chr9 | 15568341 | 15568345 | 4 | |
| Deletion | Chr9 | 16013679 | 16013688 | 9 | |
| Deletion | Chr12 | 16443635 | 16443636 | 1 | |
| Deletion | Chr10 | 17008001 | 17056000 | 48000 | 6 |
| Deletion | Chr10 | 17017169 | 17017171 | 2 | |
| Deletion | Chr3 | 17160386 | 17160395 | 9 | |
| Deletion | Chr8 | 17319001 | 17489000 | 170000 | 21 |
| Deletion | Chr10 | 17442001 | 17625000 | 183000 | 21 |
| Deletion | Chr8 | 17497001 | 17542000 | 45000 | 7 |
| Deletion | Chr8 | 17550001 | 17709000 | 159000 | 24 |
| Deletion | Chr1 | 18915877 | 18915879 | 2 | |
| Deletion | Chr8 | 20073111 | 20073115 | 4 | |
| Deletion | Chr11 | 21524078 | 21524081 | 3 | |
| Deletion | Chr12 | 21979335 | 21979388 | 53 | |
| Deletion | Chr9 | 21985852 | 21985853 | 1 | |
| Deletion | Chr12 | 22586453 | 22586455 | 2 | |
| Deletion | Chr8 | 24915172 | 24915178 | 6 | |
| Deletion | Chr1 | 25420001 | 25458000 | 38000 | 7 |
| Deletion | Chr1 | 25514001 | 25532000 | 18000 | 3 |
| Deletion | Chr1 | 25703001 | 26013000 | 310000 | 49 |
| Deletion | Chr2 | 25921001 | 25979000 | 58000 | 9 |
| Deletion | Chr1 | 26134001 | 26207000 | 73000 | 10 |
| Deletion | Chr1 | 26379274 | 26379275 | 1 | |
| Deletion | Chr1 | 26610149 | 26610150 | 1 | |
| Deletion | Chr1 | 26679001 | 26750000 | 71000 | 10 |
| Deletion | Chr4 | 29468001 | 29479000 | 11000 | LOC_Os04g49380 |
| Deletion | Chr3 | 31936839 | 31936840 | 1 | |
| Deletion | Chr2 | 32367414 | 32367420 | 6 |
Insertions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11552138 | 11552138 | 1 | LOC_Os01g20740 |
| Insertion | Chr1 | 38203564 | 38203567 | 4 | |
| Insertion | Chr12 | 26070973 | 26070973 | 1 | |
| Insertion | Chr2 | 22701818 | 22701843 | 26 | |
| Insertion | Chr2 | 22787332 | 22787332 | 1 | |
| Insertion | Chr4 | 8611649 | 8611649 | 1 | |
| Insertion | Chr5 | 12952156 | 12952156 | 1 | |
| Insertion | Chr5 | 1336836 | 1336836 | 1 | |
| Insertion | Chr5 | 8835314 | 8835314 | 1 | |
| Insertion | Chr6 | 4290180 | 4290180 | 1 | |
| Insertion | Chr8 | 102365 | 102368 | 4 | |
| Insertion | Chr8 | 9831785 | 9831785 | 1 | |
| Insertion | Chr9 | 16173455 | 16173455 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 1483449 | 1483710 | |
| Inversion | Chr9 | 16050924 | 16051105 | LOC_Os09g26550 |
| Inversion | Chr7 | 22955946 | 22956070 | LOC_Os07g38270 |
| Inversion | Chr1 | 25419484 | 25702846 | |
| Inversion | Chr1 | 39390117 | 39390326 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 14791364 | Chr1 | 25725454 | |
| Translocation | Chr9 | 14791736 | Chr8 | 17313139 | |
| Translocation | Chr8 | 24363151 | Chr2 | 1947526 | |
| Translocation | Chr5 | 27610094 | Chr1 | 25420715 | |
| Translocation | Chr5 | 27610105 | Chr1 | 25419529 | LOC_Os05g48170 |
