Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4382-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4382-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 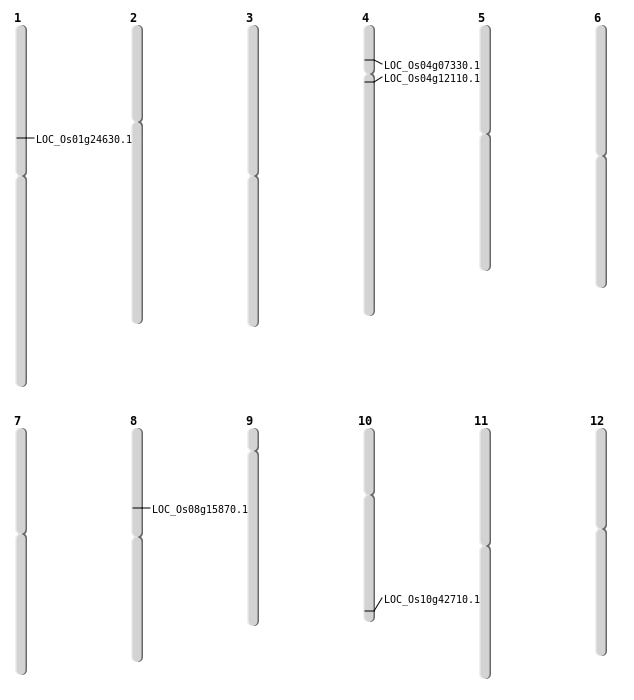
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17771774 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 34078564 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 41458085 | C-T | HET | INTRON | |
| SBS | Chr1 | 4331252 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 6550299 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 8505268 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 23037745 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g42710 |
| SBS | Chr11 | 15557894 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 2646540 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 9760715 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 6624129 | C-A | HET | INTRON | |
| SBS | Chr2 | 14257677 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 15575652 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 3408865 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr2 | 4531685 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 1355026 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 16618534 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 20457145 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 27268382 | G-A | HET | INTRON | |
| SBS | Chr3 | 5478091 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 6892611 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 3904158 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g07330 |
| SBS | Chr4 | 6660236 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g12110 |
| SBS | Chr4 | 6660237 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 6660261 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 11615586 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 22776123 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr7 | 10643327 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 11225208 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr7 | 11613180 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 17447113 | A-T | HET | INTRON |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 314092 | 314094 | 2 | |
| Deletion | Chr7 | 420607 | 420608 | 1 | |
| Deletion | Chr5 | 1221739 | 1221742 | 3 | |
| Deletion | Chr6 | 1369799 | 1369801 | 2 | |
| Deletion | Chr8 | 2488402 | 2488461 | 59 | |
| Deletion | Chr6 | 2556750 | 2556751 | 1 | |
| Deletion | Chr11 | 3001998 | 3001999 | 1 | |
| Deletion | Chr4 | 3100936 | 3100939 | 3 | |
| Deletion | Chr7 | 3146407 | 3146461 | 54 | |
| Deletion | Chr6 | 6138932 | 6138933 | 1 | |
| Deletion | Chr7 | 6154911 | 6154933 | 22 | |
| Deletion | Chr8 | 9651378 | 9651380 | 2 | LOC_Os08g15870 |
| Deletion | Chr6 | 10622549 | 10622714 | 165 | |
| Deletion | Chr12 | 11141520 | 11141604 | 84 | |
| Deletion | Chr8 | 12526087 | 12526088 | 1 | |
| Deletion | Chr7 | 13797785 | 13797786 | 1 | |
| Deletion | Chr1 | 13873512 | 13873513 | 1 | LOC_Os01g24630 |
| Deletion | Chr10 | 15257672 | 15257673 | 1 | |
| Deletion | Chr1 | 16975172 | 16975173 | 1 | |
| Deletion | Chr5 | 17562409 | 17562439 | 30 | |
| Deletion | Chr9 | 18632402 | 18632403 | 1 | |
| Deletion | Chr1 | 20243271 | 20243273 | 2 | |
| Deletion | Chr10 | 20803854 | 20803855 | 1 | |
| Deletion | Chr7 | 24223003 | 24223004 | 1 | |
| Deletion | Chr2 | 25967668 | 25967669 | 1 | |
| Deletion | Chr12 | 27027671 | 27027721 | 50 | |
| Deletion | Chr12 | 27233424 | 27233426 | 2 | |
| Deletion | Chr2 | 27351179 | 27351180 | 1 | |
| Deletion | Chr4 | 30357264 | 30357265 | 1 | |
| Deletion | Chr4 | 30841115 | 30841116 | 1 | |
| Deletion | Chr1 | 34783444 | 34783445 | 1 | |
| Deletion | Chr1 | 38207093 | 38207097 | 4 |
Insertions: 15
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 33518179 | 33518390 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 5113843 | Chr4 | 2989432 |
