Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4383-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4383-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 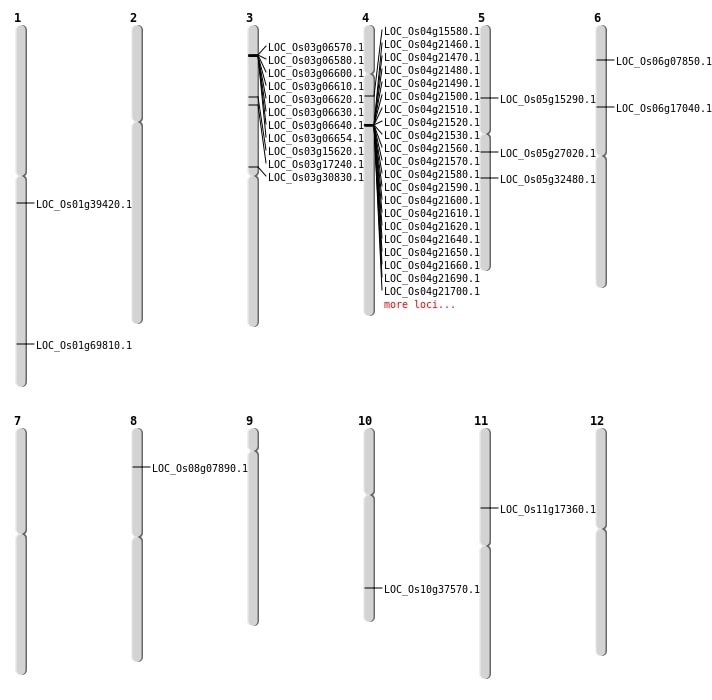
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1271085 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 13070785 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 19887329 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 20998790 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 22213372 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g39420 |
| SBS | Chr1 | 40825122 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 8279762 | A-T | HET | INTRON | |
| SBS | Chr10 | 11473176 | G-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 20109970 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g37570 |
| SBS | Chr11 | 21227142 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 9676344 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g17360 |
| SBS | Chr12 | 11641737 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 12970050 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 15047973 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 2183457 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 23011948 | T-C | HET | INTRON | |
| SBS | Chr2 | 18837655 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 6718073 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 6769724 | C-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 7275411 | A-G | HOMO | INTRON | |
| SBS | Chr3 | 14895238 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 16202631 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 17571518 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 17571519 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g30830 |
| SBS | Chr3 | 21303837 | G-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 3177011 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 3177012 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 36203646 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 8617634 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g15620 |
| SBS | Chr3 | 9685453 | A-G | HET | INTRON | |
| SBS | Chr4 | 1722144 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 9720446 | T-A | HET | INTRON | |
| SBS | Chr4 | 9720449 | G-A | HET | INTRON | |
| SBS | Chr5 | 6308021 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 8672466 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g15290 |
| SBS | Chr7 | 3614834 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 16667029 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 27832027 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 6317439 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 4734936 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 7972352 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 8163400 | G-A | HET | INTERGENIC |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 346730 | 346731 | 1 | |
| Deletion | Chr6 | 718495 | 718496 | 1 | |
| Deletion | Chr11 | 1920689 | 1920690 | 1 | |
| Deletion | Chr10 | 3272816 | 3272817 | 1 | |
| Deletion | Chr3 | 3309001 | 3369000 | 60000 | 8 |
| Deletion | Chr6 | 4833588 | 4833598 | 10 | |
| Deletion | Chr12 | 5370558 | 5370559 | 1 | |
| Deletion | Chr7 | 7137997 | 7137999 | 2 | |
| Deletion | Chr12 | 9296439 | 9296440 | 1 | |
| Deletion | Chr9 | 9965983 | 9965984 | 1 | |
| Deletion | Chr4 | 11199352 | 11199353 | 1 | |
| Deletion | Chr1 | 11489605 | 11489606 | 1 | |
| Deletion | Chr4 | 12133001 | 12260000 | 127000 | 18 |
| Deletion | Chr4 | 12271001 | 12345000 | 74000 | 12 |
| Deletion | Chr4 | 12910001 | 13107000 | 197000 | 28 |
| Deletion | Chr10 | 15136621 | 15136632 | 11 | |
| Deletion | Chr8 | 15504428 | 15504429 | 1 | |
| Deletion | Chr12 | 15540968 | 15540969 | 1 | |
| Deletion | Chr5 | 15712436 | 15712457 | 21 | LOC_Os05g27020 |
| Deletion | Chr9 | 16167452 | 16167453 | 1 | |
| Deletion | Chr7 | 16285725 | 16285731 | 6 | |
| Deletion | Chr12 | 16581631 | 16581633 | 2 | |
| Deletion | Chr9 | 17711843 | 17711845 | 2 | |
| Deletion | Chr5 | 18974415 | 18974443 | 28 | LOC_Os05g32480 |
| Deletion | Chr2 | 23475392 | 23475481 | 89 | |
| Deletion | Chr3 | 23658353 | 23658362 | 9 | |
| Deletion | Chr4 | 25943301 | 25943302 | 1 | |
| Deletion | Chr7 | 27685037 | 27685038 | 1 | |
| Deletion | Chr6 | 29130256 | 29130263 | 7 | |
| Deletion | Chr6 | 29155834 | 29155840 | 6 | |
| Deletion | Chr4 | 30753349 | 30753350 | 1 | |
| Deletion | Chr1 | 32690993 | 32690994 | 1 | |
| Deletion | Chr1 | 38757171 | 38757172 | 1 | |
| Deletion | Chr1 | 39989989 | 39989991 | 2 |
Insertions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 40308970 | 40308970 | 1 | LOC_Os01g69810 |
| Insertion | Chr10 | 16148386 | 16148386 | 1 | |
| Insertion | Chr10 | 2478001 | 2478037 | 37 | |
| Insertion | Chr10 | 3591300 | 3591334 | 35 | |
| Insertion | Chr11 | 14505114 | 14505114 | 1 | |
| Insertion | Chr11 | 8175197 | 8175202 | 6 | |
| Insertion | Chr2 | 18676048 | 18676048 | 1 | |
| Insertion | Chr2 | 23462455 | 23462455 | 1 | |
| Insertion | Chr3 | 7732517 | 7732517 | 1 | |
| Insertion | Chr5 | 10754448 | 10754501 | 54 | |
| Insertion | Chr5 | 23126717 | 23126718 | 2 | |
| Insertion | Chr6 | 3819118 | 3819118 | 1 | LOC_Os06g07850 |
| Insertion | Chr6 | 4830624 | 4830624 | 1 | |
| Insertion | Chr6 | 9882072 | 9882078 | 7 | LOC_Os06g17040 |
| Insertion | Chr7 | 12585262 | 12585262 | 1 | |
| Insertion | Chr8 | 9136215 | 9136220 | 6 | |
| Insertion | Chr9 | 3012431 | 3012431 | 1 | |
| Insertion | Chr9 | 7017568 | 7017568 | 1 |
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 8472680 | 8472836 | LOC_Os04g15580 |
| Inversion | Chr5 | 9589898 | 9590025 | |
| Inversion | Chr11 | 10983349 | 10983453 | |
| Inversion | Chr10 | 13826572 | 13826716 | |
| Inversion | Chr1 | 16539575 | 16539884 | |
| Inversion | Chr1 | 30207673 | 30208102 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 4443487 | Chr8 | 15117194 | LOC_Os08g07890 |
| Translocation | Chr12 | 4447529 | Chr4 | 27074184 | |
| Translocation | Chr4 | 9605634 | Chr3 | 3307188 | LOC_Os03g17240 |
| Translocation | Chr5 | 16313876 | Chr3 | 3306349 | |
| Translocation | Chr4 | 27074179 | Chr2 | 33553003 |
