Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4387-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4387-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 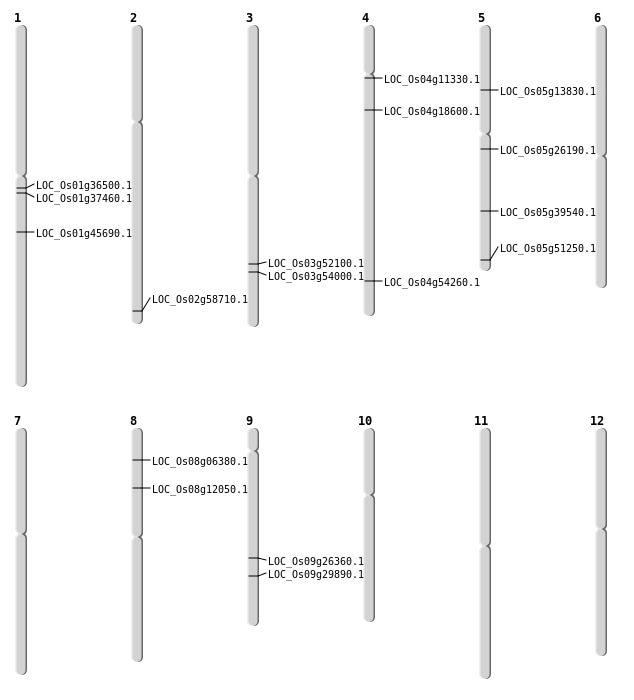
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20259582 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g36500 |
| SBS | Chr1 | 22974517 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 24362495 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 25943273 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g45690 |
| SBS | Chr1 | 5386586 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 12386668 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 3132179 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 21440128 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 20392717 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 24943531 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr12 | 4003025 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 12054792 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 14660365 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 19355343 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 35867711 | C-T | HET | STOP_GAINED | LOC_Os02g58710 |
| SBS | Chr2 | 7092735 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 14763819 | C-A | HET | INTRON | |
| SBS | Chr3 | 29107467 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 29914889 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g52100 |
| SBS | Chr4 | 10279765 | T-C | HET | STOP_LOST | LOC_Os04g18600 |
| SBS | Chr4 | 23438583 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 29335956 | C-T | HET | INTRON | |
| SBS | Chr4 | 6184658 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g11330 |
| SBS | Chr4 | 9700372 | C-T | HET | INTRON | |
| SBS | Chr5 | 10162662 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 20195354 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 21340022 | G-C | HET | INTRON | |
| SBS | Chr5 | 26040563 | T-C | HET | INTRON | |
| SBS | Chr5 | 28762454 | T-C | HET | INTRON | |
| SBS | Chr5 | 7685773 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g13830 |
| SBS | Chr6 | 173772 | C-T | HET | INTRON | |
| SBS | Chr6 | 17407980 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 23754804 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 6074044 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 7304589 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 18499260 | T-C | HET | INTRON | |
| SBS | Chr7 | 28917145 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 5502373 | T-C | HET | INTRON | |
| SBS | Chr8 | 15977059 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr8 | 19328382 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 22451583 | T-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 3545309 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g06380 |
| SBS | Chr8 | 7095143 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g12050 |
| SBS | Chr9 | 15727775 | C-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 20564720 | A-T | HOMO | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 176985 | 177002 | 17 | |
| Deletion | Chr3 | 1037586 | 1037595 | 9 | |
| Deletion | Chr3 | 2253271 | 2253275 | 4 | |
| Deletion | Chr3 | 4645325 | 4645328 | 3 | |
| Deletion | Chr1 | 5674041 | 5674042 | 1 | |
| Deletion | Chr3 | 12044835 | 12044839 | 4 | |
| Deletion | Chr4 | 13306934 | 13306935 | 1 | |
| Deletion | Chr9 | 13577000 | 13577001 | 1 | |
| Deletion | Chr5 | 15240652 | 15240654 | 2 | LOC_Os05g26190 |
| Deletion | Chr6 | 17957574 | 17957577 | 3 | |
| Deletion | Chr9 | 18175082 | 18175083 | 1 | LOC_Os09g29890 |
| Deletion | Chr9 | 18817214 | 18817215 | 1 | |
| Deletion | Chr12 | 19238706 | 19238708 | 2 | |
| Deletion | Chr7 | 20498435 | 20498436 | 1 | |
| Deletion | Chr2 | 20747531 | 20747532 | 1 | |
| Deletion | Chr3 | 20852136 | 20852137 | 1 | |
| Deletion | Chr1 | 20936978 | 20937010 | 32 | LOC_Os01g37460 |
| Deletion | Chr11 | 25899558 | 25899559 | 1 | |
| Deletion | Chr4 | 25996161 | 25996162 | 1 | |
| Deletion | Chr2 | 26567035 | 26567114 | 79 | |
| Deletion | Chr7 | 27412388 | 27412389 | 1 | |
| Deletion | Chr5 | 29396274 | 29396283 | 9 | LOC_Os05g51250 |
| Deletion | Chr3 | 30299257 | 30299258 | 1 | |
| Deletion | Chr3 | 30961268 | 30961270 | 2 | LOC_Os03g54000 |
| Deletion | Chr4 | 32308896 | 32308897 | 1 | LOC_Os04g54260 |
| Deletion | Chr1 | 38844018 | 38844027 | 9 | |
| Deletion | Chr1 | 41710165 | 41710167 | 2 |
Insertions: 17
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 2742627 | 3689578 | |
| Inversion | Chr3 | 2742641 | 3689584 | |
| Inversion | Chr4 | 12674012 | 12674213 | |
| Inversion | Chr9 | 15921728 | 16263895 | LOC_Os09g26360 |
| Inversion | Chr9 | 15921731 | 16264034 | 2 |
| Inversion | Chr9 | 19759068 | 19759249 | |
| Inversion | Chr6 | 20298466 | 20298637 | |
| Inversion | Chr5 | 22781431 | 23204668 | LOC_Os05g39540 |
No Translocation
