Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN439-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN439-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 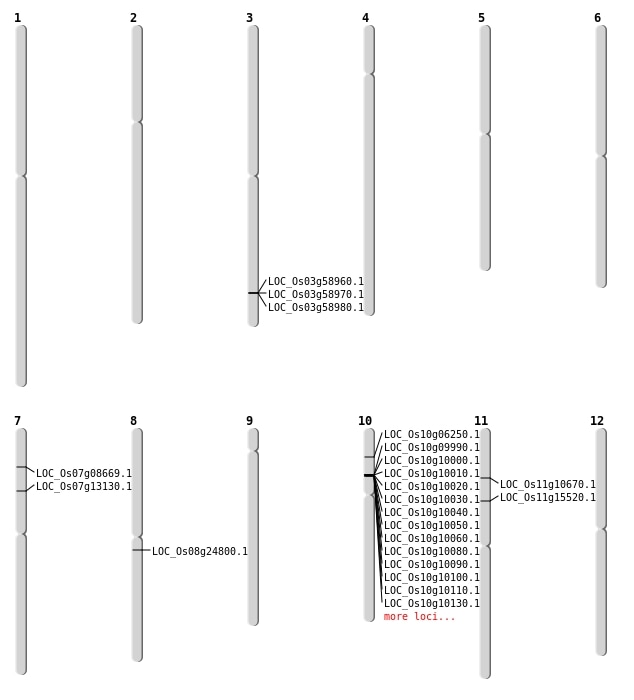
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12359186 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 13349971 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 33748671 | G-A | HET | INTRON | |
| SBS | Chr1 | 34127679 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 42562725 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 11150916 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 22552254 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 3206804 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g06250 |
| SBS | Chr11 | 10281484 | C-T | HET | INTRON | |
| SBS | Chr11 | 10399675 | C-T | HET | INTRON | |
| SBS | Chr11 | 11917951 | T-G | HOMO | INTRON | |
| SBS | Chr11 | 12293419 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 15677423 | T-A | HET | INTRON | |
| SBS | Chr11 | 16666643 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 22325913 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 15373126 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 18901192 | C-A | HET | INTRON | |
| SBS | Chr2 | 32840304 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 18818890 | C-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 2327392 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 27249797 | T-C | HOMO | INTRON | |
| SBS | Chr3 | 5024961 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 27437110 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 697838 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 23941935 | C-T | HET | INTRON | |
| SBS | Chr6 | 19200316 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 20396927 | A-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 28840436 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 12745081 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 14519260 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 2458484 | T-G | HOMO | UTR_3_PRIME | |
| SBS | Chr7 | 7296632 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 7515798 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g13130 |
| SBS | Chr7 | 8871409 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 11002016 | G-A | HOMO | INTERGENIC |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 139807 | 139809 | 2 | |
| Deletion | Chr4 | 665927 | 665931 | 4 | |
| Deletion | Chr10 | 776425 | 776434 | 9 | |
| Deletion | Chr11 | 1052158 | 1052169 | 11 | |
| Deletion | Chr7 | 4467472 | 4467478 | 6 | LOC_Os07g08669 |
| Deletion | Chr10 | 5412001 | 5571000 | 159000 | 16 |
| Deletion | Chr9 | 5669826 | 5669849 | 23 | |
| Deletion | Chr11 | 5852150 | 5852156 | 6 | LOC_Os11g10670 |
| Deletion | Chr10 | 6183773 | 6183775 | 2 | |
| Deletion | Chr6 | 8269361 | 8269370 | 9 | |
| Deletion | Chr11 | 8795589 | 8795595 | 6 | LOC_Os11g15520 |
| Deletion | Chr11 | 9687927 | 9687928 | 1 | |
| Deletion | Chr10 | 10404309 | 10404320 | 11 | |
| Deletion | Chr12 | 10771479 | 10771495 | 16 | |
| Deletion | Chr8 | 11836891 | 11836892 | 1 | |
| Deletion | Chr9 | 12237090 | 12237096 | 6 | |
| Deletion | Chr6 | 12480483 | 12480489 | 6 | |
| Deletion | Chr7 | 13108568 | 13108577 | 9 | |
| Deletion | Chr3 | 14525271 | 14525272 | 1 | |
| Deletion | Chr7 | 16399552 | 16399557 | 5 | |
| Deletion | Chr10 | 19790715 | 19790716 | 1 | |
| Deletion | Chr3 | 21097289 | 21097290 | 1 | |
| Deletion | Chr7 | 22334892 | 22334908 | 16 | |
| Deletion | Chr9 | 22561816 | 22561825 | 9 | |
| Deletion | Chr7 | 22951805 | 22951810 | 5 | |
| Deletion | Chr8 | 23606308 | 23606310 | 2 | |
| Deletion | Chr12 | 24093262 | 24093285 | 23 | |
| Deletion | Chr3 | 24148502 | 24148503 | 1 | |
| Deletion | Chr6 | 26403531 | 26403532 | 1 | |
| Deletion | Chr1 | 26640564 | 26640566 | 2 | |
| Deletion | Chr1 | 27037620 | 27037622 | 2 | |
| Deletion | Chr4 | 29992398 | 29992399 | 1 | |
| Deletion | Chr3 | 33570773 | 33588228 | 17455 | 3 |
| Deletion | Chr3 | 36047656 | 36047657 | 1 |
Insertions: 6
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 5348812 | 5377542 | |
| Inversion | Chr10 | 5348823 | 5570635 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 15011896 | Chr6 | 25504578 | LOC_Os08g24800 |
