Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4392-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4392-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 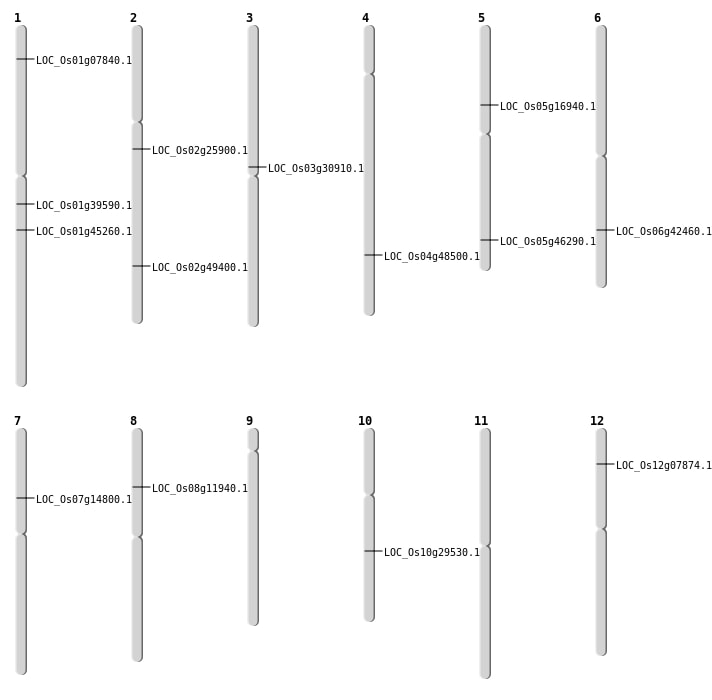
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22332783 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g39590 |
| SBS | Chr1 | 25580344 | T-A | HOMO | INTRON | |
| SBS | Chr1 | 27326851 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 3772730 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g07840 |
| SBS | Chr1 | 42384056 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 13817204 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 15340528 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g29530 |
| SBS | Chr10 | 20993088 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 22720125 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 8395786 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 10316586 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 16694603 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 3985629 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g07874 |
| SBS | Chr2 | 10272734 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 15194992 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g25900 |
| SBS | Chr2 | 15263694 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 2050914 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 28365480 | G-A | HET | INTRON | |
| SBS | Chr3 | 17590829 | T-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 21031127 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr4 | 21629096 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 28919700 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g48500 |
| SBS | Chr4 | 34478227 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 25113116 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 9657223 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g16940 |
| SBS | Chr6 | 16951031 | C-T | HET | INTRON | |
| SBS | Chr6 | 20035147 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr6 | 25521816 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g42460 |
| SBS | Chr7 | 21112242 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 21547209 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 23355938 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 24789748 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 9936647 | G-A | HET | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 1843385 | 1843386 | 1 | |
| Deletion | Chr8 | 3097620 | 3097621 | 1 | |
| Deletion | Chr9 | 3288363 | 3288365 | 2 | |
| Deletion | Chr5 | 3860008 | 3860012 | 4 | |
| Deletion | Chr7 | 8449006 | 8449007 | 1 | LOC_Os07g14800 |
| Deletion | Chr11 | 9907912 | 9907913 | 1 | |
| Deletion | Chr2 | 12196912 | 12196913 | 1 | |
| Deletion | Chr4 | 14096205 | 14096206 | 1 | |
| Deletion | Chr12 | 14285588 | 14285596 | 8 | |
| Deletion | Chr9 | 15685627 | 15685628 | 1 | |
| Deletion | Chr3 | 16650211 | 16650234 | 23 | |
| Deletion | Chr2 | 17363398 | 17363400 | 2 | |
| Deletion | Chr3 | 17607586 | 17607600 | 14 | LOC_Os03g30910 |
| Deletion | Chr12 | 18184620 | 18184622 | 2 | |
| Deletion | Chr2 | 19416092 | 19416097 | 5 | |
| Deletion | Chr6 | 19581469 | 19581470 | 1 | |
| Deletion | Chr5 | 19704469 | 19704470 | 1 | |
| Deletion | Chr7 | 24347182 | 24347223 | 41 | |
| Deletion | Chr1 | 24945307 | 24945309 | 2 | |
| Deletion | Chr1 | 25690741 | 25690742 | 1 | LOC_Os01g45260 |
| Deletion | Chr4 | 27597878 | 27597879 | 1 | |
| Deletion | Chr3 | 28994603 | 28994611 | 8 | |
| Deletion | Chr2 | 30190701 | 30190703 | 2 | LOC_Os02g49400 |
| Deletion | Chr1 | 33754010 | 33754016 | 6 | |
| Deletion | Chr2 | 34380134 | 34380135 | 1 |
Insertions: 21
Inversions: 9
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 9511394 | 10113434 | |
| Inversion | Chr6 | 10839767 | 10839942 | |
| Inversion | Chr10 | 16730186 | 16730340 | |
| Inversion | Chr10 | 18787308 | 18912457 | |
| Inversion | Chr10 | 18787312 | 18912457 | |
| Inversion | Chr11 | 25259214 | 25259387 | |
| Inversion | Chr5 | 26836235 | 26836537 | LOC_Os05g46290 |
| Inversion | Chr3 | 29904412 | 29904532 | |
| Inversion | Chr1 | 35883165 | 35883287 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 7002688 | Chr3 | 13461614 | LOC_Os08g11940 |
| Translocation | Chr10 | 18760551 | Chr6 | 9511388 | |
| Translocation | Chr10 | 18760707 | Chr6 | 10113428 | |
| Translocation | Chr10 | 19026764 | Chr3 | 17936656 |
