Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4394-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4394-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 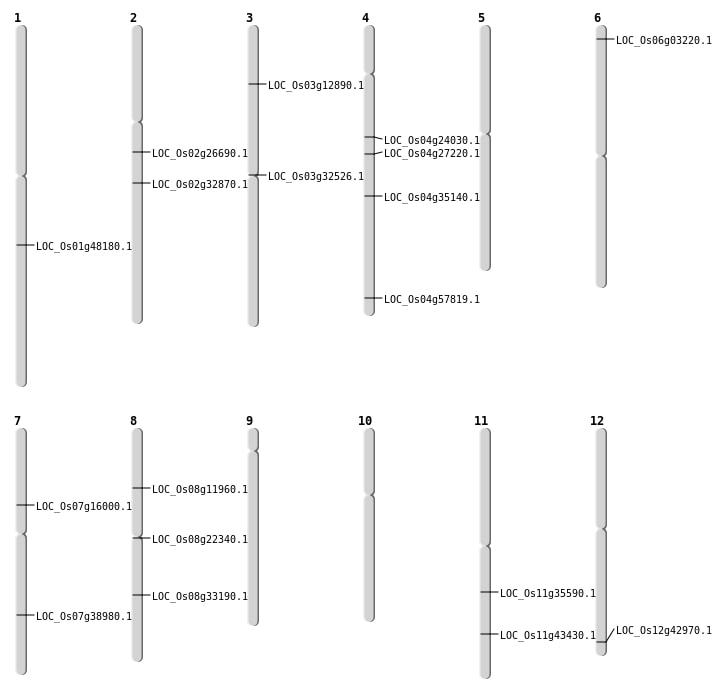
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11608410 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 17228204 | A-C | HOMO | INTRON | |
| SBS | Chr1 | 25170912 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 2583944 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 6587977 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 8903091 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 1874792 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 20269047 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 21936171 | G-A | HET | INTRON | |
| SBS | Chr11 | 26078623 | T-C | HOMO | INTRON | |
| SBS | Chr11 | 4522944 | C-T | HET | INTRON | |
| SBS | Chr11 | 9358500 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 21729387 | A-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 21729388 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 26695303 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g42970 |
| SBS | Chr2 | 19522074 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g32870 |
| SBS | Chr3 | 1002554 | G-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 1869299 | G-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 20481308 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 25931195 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 29868474 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 32324699 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 1071928 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 13730730 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g24030 |
| SBS | Chr4 | 1396653 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 29943957 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 3248645 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 35312053 | A-G | HOMO | INTRON | |
| SBS | Chr5 | 13681409 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 24505677 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 24859375 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 6226998 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 646898 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 8010833 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 9377793 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 12939076 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 23366977 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g38980 |
| SBS | Chr7 | 8454097 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 9323629 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g16000 |
| SBS | Chr8 | 10386304 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 11298628 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 23126818 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 7009647 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g11960 |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 1219043 | 1219061 | 18 | LOC_Os06g03220 |
| Deletion | Chr2 | 1498671 | 1498672 | 1 | |
| Deletion | Chr5 | 4555505 | 4555506 | 1 | |
| Deletion | Chr5 | 5570951 | 5570952 | 1 | |
| Deletion | Chr3 | 6948267 | 6948272 | 5 | LOC_Os03g12890 |
| Deletion | Chr1 | 9013496 | 9013498 | 2 | |
| Deletion | Chr7 | 9372390 | 9372393 | 3 | |
| Deletion | Chr8 | 13462575 | 13462576 | 1 | LOC_Os08g22340 |
| Deletion | Chr2 | 15671581 | 15671582 | 1 | LOC_Os02g26690 |
| Deletion | Chr4 | 16087423 | 16087425 | 2 | LOC_Os04g27220 |
| Deletion | Chr7 | 17081889 | 17081899 | 10 | |
| Deletion | Chr9 | 18206709 | 18206710 | 1 | |
| Deletion | Chr3 | 18604276 | 18604285 | 9 | LOC_Os03g32526 |
| Deletion | Chr3 | 18770641 | 18770642 | 1 | |
| Deletion | Chr8 | 18772457 | 18772458 | 1 | |
| Deletion | Chr2 | 18855217 | 18855218 | 1 | |
| Deletion | Chr6 | 20069015 | 20069041 | 26 | |
| Deletion | Chr8 | 20654895 | 20654896 | 1 | LOC_Os08g33190 |
| Deletion | Chr2 | 20743406 | 20743407 | 1 | |
| Deletion | Chr11 | 20871930 | 20871931 | 1 | LOC_Os11g35590 |
| Deletion | Chr4 | 21360311 | 21360312 | 1 | LOC_Os04g35140 |
| Deletion | Chr11 | 26230358 | 26230364 | 6 | LOC_Os11g43430 |
| Deletion | Chr6 | 27081999 | 27082009 | 10 | |
| Deletion | Chr1 | 27597397 | 27597410 | 13 | LOC_Os01g48180 |
| Deletion | Chr6 | 27724420 | 27724445 | 25 | |
| Deletion | Chr3 | 28954799 | 28954804 | 5 | |
| Deletion | Chr5 | 29586789 | 29586790 | 1 | |
| Deletion | Chr2 | 29700174 | 29700175 | 1 | |
| Deletion | Chr2 | 29811248 | 29811249 | 1 |
Insertions: 9
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 4421483 | 4421608 | |
| Inversion | Chr6 | 4710368 | 4710534 | |
| Inversion | Chr7 | 10928855 | 10928948 | |
| Inversion | Chr6 | 11408006 | 11408167 | |
| Inversion | Chr11 | 26487512 | 26487721 | |
| Inversion | Chr4 | 34431329 | 34431513 | LOC_Os04g57819 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 4210432 | Chr2 | 12350483 | |
| Translocation | Chr5 | 4210437 | Chr2 | 12352422 |
