Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4396-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4396-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 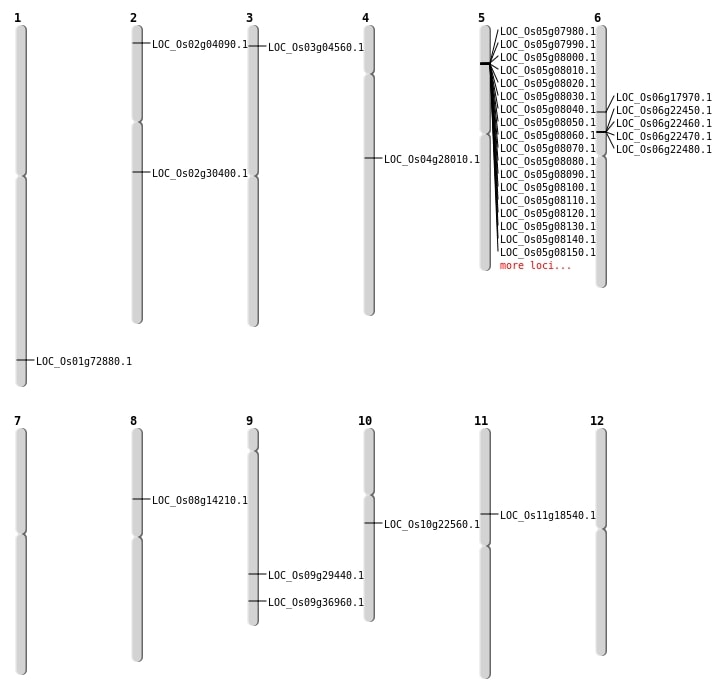
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 32499036 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 42273127 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g72880 |
| SBS | Chr10 | 11691754 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g22560 |
| SBS | Chr10 | 14362515 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 19138527 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 13202825 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 13069720 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 1592471 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 24641398 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 8870722 | A-G | HET | INTRON | |
| SBS | Chr2 | 16623525 | A-G | HOMO | INTRON | |
| SBS | Chr2 | 1780449 | A-T | HET | STOP_GAINED | LOC_Os02g04090 |
| SBS | Chr2 | 19267032 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 2128025 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g04560 |
| SBS | Chr3 | 4917767 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 4917768 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 13851625 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 18080981 | C-T | HET | INTRON | |
| SBS | Chr4 | 28698778 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 11435151 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr6 | 10389418 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 11987760 | A-G | HOMO | INTRON | |
| SBS | Chr6 | 26716162 | T-A | HET | INTRON | |
| SBS | Chr6 | 30158103 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 6054750 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 4669822 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 21172918 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 8554232 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 17893042 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g29440 |
| SBS | Chr9 | 21516437 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 2980035 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 9705958 | C-T | HET | INTRON | |
| SBS | Chr9 | 9763222 | A-G | HET | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 3785160 | 3785162 | 2 | |
| Deletion | Chr5 | 4319001 | 4609000 | 290000 | 47 |
| Deletion | Chr5 | 4515065 | 4515066 | 1 | |
| Deletion | Chr5 | 5669206 | 5669221 | 15 | |
| Deletion | Chr11 | 7153733 | 7153734 | 1 | |
| Deletion | Chr2 | 10185620 | 10185634 | 14 | |
| Deletion | Chr12 | 11689736 | 11689738 | 2 | |
| Deletion | Chr6 | 13048001 | 13067000 | 19000 | 4 |
| Deletion | Chr12 | 15947136 | 15947137 | 1 | |
| Deletion | Chr12 | 16443050 | 16443053 | 3 | |
| Deletion | Chr12 | 16843520 | 16843529 | 9 | |
| Deletion | Chr2 | 18107719 | 18107725 | 6 | LOC_Os02g30400 |
| Deletion | Chr1 | 19900788 | 19900789 | 1 | |
| Deletion | Chr2 | 21601923 | 21601929 | 6 | |
| Deletion | Chr5 | 22139600 | 22139602 | 2 | |
| Deletion | Chr9 | 22456585 | 22456597 | 12 | |
| Deletion | Chr2 | 22902795 | 22902796 | 1 | |
| Deletion | Chr1 | 23018242 | 23018245 | 3 | |
| Deletion | Chr12 | 23068772 | 23068773 | 1 | |
| Deletion | Chr2 | 26017759 | 26017760 | 1 | |
| Deletion | Chr8 | 26887078 | 26887084 | 6 | |
| Deletion | Chr11 | 28909080 | 28909085 | 5 | |
| Deletion | Chr4 | 29209202 | 29209246 | 44 | |
| Deletion | Chr3 | 31100296 | 31100297 | 1 | |
| Deletion | Chr3 | 31594071 | 31594072 | 1 | |
| Deletion | Chr1 | 35934460 | 35934461 | 1 | |
| Deletion | Chr1 | 40412956 | 40412957 | 1 |
Insertions: 13
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 14670197 | 14670344 | |
| Inversion | Chr4 | 16540731 | 16540875 | LOC_Os04g28010 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 8494271 | Chr1 | 32071121 | LOC_Os08g14210 |
| Translocation | Chr11 | 10449196 | Chr6 | 17476301 | 2 |
| Translocation | Chr12 | 18596757 | Chr5 | 10270578 | |
| Translocation | Chr9 | 21320883 | Chr7 | 16054069 | LOC_Os09g36960 |
| Translocation | Chr9 | 21578730 | Chr7 | 16054069 |
