Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4397-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4397-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 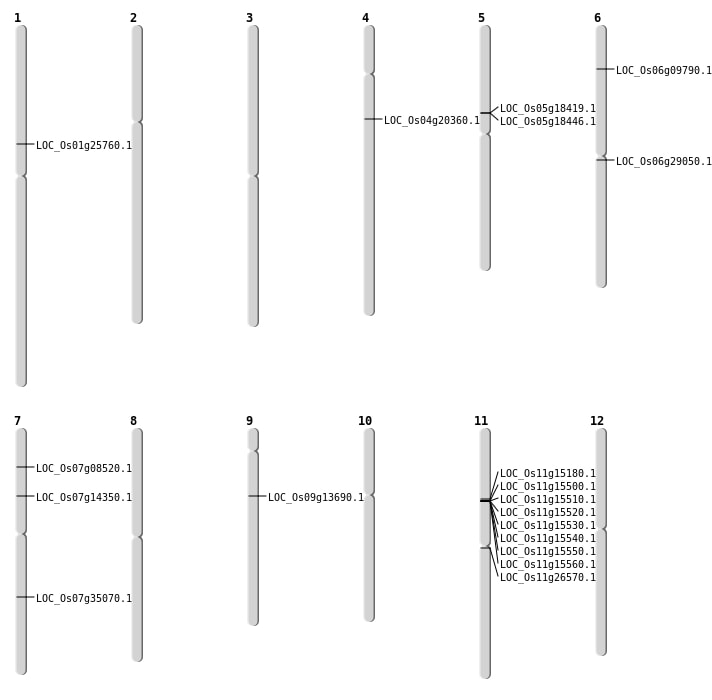
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14291571 | G-T | HET | UTR_3_PRIME | |
| SBS | Chr1 | 36742930 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 42013362 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 7624833 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 8744327 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 9923333 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 18863651 | C-T | HET | INTRON | |
| SBS | Chr11 | 11279044 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 15224211 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g26570 |
| SBS | Chr11 | 23504649 | C-A | HET | INTRON | |
| SBS | Chr11 | 23504650 | T-A | HET | INTRON | |
| SBS | Chr11 | 8268882 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 15613360 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 10905833 | G-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 34745276 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 35662534 | A-G | HET | INTRON | |
| SBS | Chr3 | 27781575 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 28287959 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 31092266 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 32468405 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 14075131 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 21380811 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 28526242 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 21008448 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g35070 |
| SBS | Chr7 | 21998340 | G-T | HET | INTRON | |
| SBS | Chr7 | 4546827 | T-C | HET | INTRON | |
| SBS | Chr7 | 7844806 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 9156866 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 13958994 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 22226335 | A-G | HOMO | UTR_5_PRIME | |
| SBS | Chr9 | 7416248 | T-C | HET | INTERGENIC |
Deletions: 33
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 701639 | 701641 | 2 | |
| Deletion | Chr8 | 2103867 | 2103897 | 30 | |
| Deletion | Chr8 | 2710572 | 2710573 | 1 | |
| Deletion | Chr6 | 4984001 | 4993000 | 9000 | LOC_Os06g09790 |
| Deletion | Chr4 | 5671722 | 5671723 | 1 | |
| Deletion | Chr3 | 6005562 | 6005567 | 5 | |
| Deletion | Chr9 | 6584917 | 6584925 | 8 | |
| Deletion | Chr6 | 7229506 | 7229511 | 5 | |
| Deletion | Chr11 | 7453124 | 7453131 | 7 | |
| Deletion | Chr9 | 8005742 | 8005749 | 7 | LOC_Os09g13690 |
| Deletion | Chr7 | 8194973 | 8194974 | 1 | LOC_Os07g14350 |
| Deletion | Chr11 | 8782001 | 8819000 | 37000 | 7 |
| Deletion | Chr2 | 9812718 | 9812719 | 1 | |
| Deletion | Chr5 | 10645001 | 10670000 | 25000 | 2 |
| Deletion | Chr4 | 11396870 | 11396873 | 3 | LOC_Os04g20360 |
| Deletion | Chr2 | 11908592 | 11908593 | 1 | |
| Deletion | Chr9 | 11983470 | 11983484 | 14 | |
| Deletion | Chr6 | 12788383 | 12788401 | 18 | |
| Deletion | Chr8 | 13096380 | 13096381 | 1 | |
| Deletion | Chr7 | 13236513 | 13236590 | 77 | |
| Deletion | Chr9 | 14291940 | 14291941 | 1 | |
| Deletion | Chr5 | 14619107 | 14619111 | 4 | |
| Deletion | Chr6 | 16556248 | 16556253 | 5 | LOC_Os06g29050 |
| Deletion | Chr7 | 17475817 | 17475819 | 2 | |
| Deletion | Chr1 | 17774692 | 17774693 | 1 | |
| Deletion | Chr3 | 18887982 | 18887991 | 9 | |
| Deletion | Chr6 | 20804683 | 20804685 | 2 | |
| Deletion | Chr4 | 22303231 | 22303240 | 9 | |
| Deletion | Chr1 | 24014217 | 24014222 | 5 | |
| Deletion | Chr11 | 25575338 | 25575340 | 2 | |
| Deletion | Chr11 | 26997635 | 26997637 | 2 | |
| Deletion | Chr4 | 30605011 | 30605028 | 17 | |
| Deletion | Chr3 | 34196951 | 34196953 | 2 |
Insertions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 22551172 | 22551172 | 1 | |
| Insertion | Chr1 | 35974754 | 35974755 | 2 | |
| Insertion | Chr10 | 16406712 | 16406713 | 2 | |
| Insertion | Chr11 | 22777217 | 22777220 | 4 | |
| Insertion | Chr11 | 8563818 | 8563828 | 11 | LOC_Os11g15180 |
| Insertion | Chr12 | 6416744 | 6416746 | 3 | |
| Insertion | Chr12 | 6707532 | 6707532 | 1 | |
| Insertion | Chr12 | 9666591 | 9666634 | 44 | |
| Insertion | Chr12 | 983508 | 983509 | 2 | |
| Insertion | Chr4 | 25995558 | 25995558 | 1 | |
| Insertion | Chr4 | 33808242 | 33808243 | 2 | |
| Insertion | Chr6 | 18410196 | 18410196 | 1 | |
| Insertion | Chr7 | 4126211 | 4126211 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 2848231 | 2848331 | |
| Inversion | Chr11 | 7260833 | 8126709 | |
| Inversion | Chr1 | 14600315 | 15470016 | LOC_Os01g25760 |
| Inversion | Chr1 | 14600790 | 15470023 | LOC_Os01g25760 |
| Inversion | Chr11 | 14960380 | 14960566 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 4402712 | Chr6 | 4993471 | LOC_Os07g08520 |
| Translocation | Chr7 | 4402717 | Chr6 | 4983830 | LOC_Os07g08520 |
| Translocation | Chr11 | 21326764 | Chr7 | 15710925 |
