Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4401-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4401-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 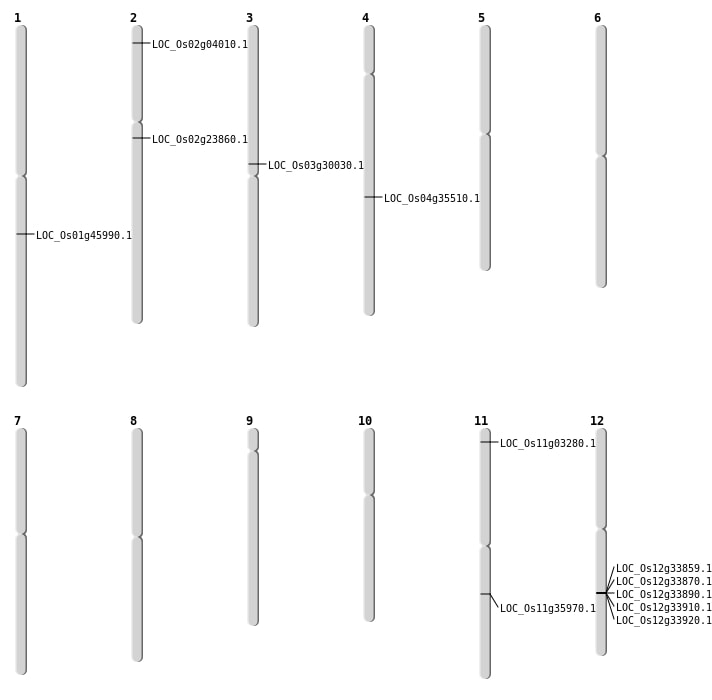
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1207003 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 26120408 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g45990 |
| SBS | Chr1 | 26579781 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 11020427 | A-C | HET | INTERGENIC | |
| SBS | Chr10 | 12213495 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 8041387 | A-T | HET | INTRON | |
| SBS | Chr11 | 16030118 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 21136238 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g35970 |
| SBS | Chr11 | 26874682 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 11876717 | C-T | HET | INTRON | |
| SBS | Chr12 | 1620452 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr12 | 24060093 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 5202403 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 9525389 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 26467434 | G-T | HET | INTRON | |
| SBS | Chr2 | 34566539 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 13502297 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 1532356 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 5587147 | A-C | HET | INTRON | |
| SBS | Chr4 | 10346746 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 22072944 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 33341955 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 8897447 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 16880422 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 28468440 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 17149679 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 21205916 | C-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 29172703 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 17505771 | T-C | HOMO | INTRON | |
| SBS | Chr7 | 3305423 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 8586838 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 1616103 | T-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 10247553 | C-A | HET | INTERGENIC |
Deletions: 36
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 3660461 | 3660462 | 1 | |
| Deletion | Chr5 | 4421241 | 4421242 | 1 | |
| Deletion | Chr5 | 7574432 | 7574477 | 45 | |
| Deletion | Chr1 | 8615204 | 8615205 | 1 | |
| Deletion | Chr9 | 9051376 | 9051388 | 12 | |
| Deletion | Chr4 | 10044865 | 10044867 | 2 | |
| Deletion | Chr11 | 11932118 | 11932139 | 21 | |
| Deletion | Chr3 | 13414166 | 13414241 | 75 | |
| Deletion | Chr9 | 13698597 | 13698598 | 1 | |
| Deletion | Chr2 | 13814995 | 13814997 | 2 | LOC_Os02g23860 |
| Deletion | Chr3 | 14101064 | 14101065 | 1 | |
| Deletion | Chr12 | 14211534 | 14211552 | 18 | |
| Deletion | Chr6 | 15284637 | 15284638 | 1 | |
| Deletion | Chr1 | 15397324 | 15397325 | 1 | |
| Deletion | Chr9 | 15428300 | 15428302 | 2 | |
| Deletion | Chr4 | 16223139 | 16223140 | 1 | |
| Deletion | Chr12 | 16750297 | 16750298 | 1 | |
| Deletion | Chr3 | 17135542 | 17135543 | 1 | LOC_Os03g30030 |
| Deletion | Chr5 | 17910567 | 17910568 | 1 | |
| Deletion | Chr4 | 19627132 | 19627133 | 1 | |
| Deletion | Chr4 | 19702505 | 19702506 | 1 | |
| Deletion | Chr5 | 20209240 | 20209241 | 1 | |
| Deletion | Chr12 | 20411001 | 20428000 | 17000 | 2 |
| Deletion | Chr12 | 20431001 | 20452000 | 21000 | 3 |
| Deletion | Chr12 | 20440542 | 20440544 | 2 | |
| Deletion | Chr12 | 20449503 | 20449505 | 2 | |
| Deletion | Chr12 | 20710669 | 20710713 | 44 | |
| Deletion | Chr6 | 21206741 | 21206747 | 6 | |
| Deletion | Chr2 | 22151521 | 22151540 | 19 | |
| Deletion | Chr2 | 22554403 | 22554405 | 2 | |
| Deletion | Chr1 | 25246729 | 25246730 | 1 | |
| Deletion | Chr1 | 25924222 | 25924223 | 1 | |
| Deletion | Chr3 | 26158345 | 26158346 | 1 | |
| Deletion | Chr6 | 26349837 | 26349838 | 1 | |
| Deletion | Chr6 | 27334067 | 27334076 | 9 | |
| Deletion | Chr3 | 30823677 | 30823689 | 12 |
Insertions: 11
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 1212871 | 1213106 | LOC_Os11g03280 |
| Inversion | Chr2 | 1736261 | 1736458 | LOC_Os02g04010 |
| Inversion | Chr4 | 21610146 | 21610350 | LOC_Os04g35510 |
| Inversion | Chr1 | 38817796 | 38817883 |
No Translocation
