Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4402-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4402-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 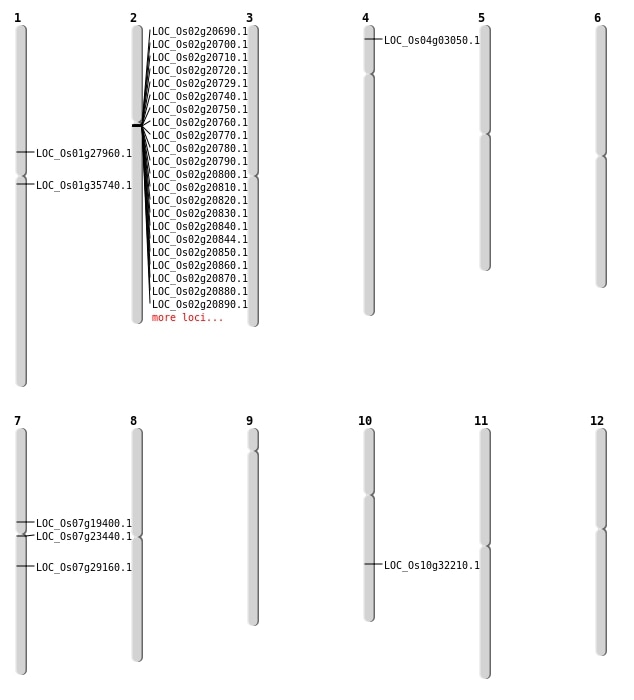
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10828802 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 19800205 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g35740 |
| SBS | Chr1 | 21710042 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 27137993 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 27539058 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 10262985 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 12002323 | A-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 16919510 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g32210 |
| SBS | Chr10 | 18246793 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 2290641 | A-T | HET | INTRON | |
| SBS | Chr10 | 943477 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 13169618 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 24302524 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 461388 | A-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 7556116 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 1238661 | G-A | HET | INTRON | |
| SBS | Chr2 | 15724248 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g26780 |
| SBS | Chr2 | 17601327 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 14247418 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 19747713 | T-C | HOMO | INTRON | |
| SBS | Chr3 | 28013903 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 30231722 | C-A | HET | INTRON | |
| SBS | Chr3 | 32312070 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 1260008 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g03050 |
| SBS | Chr4 | 2405249 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 33349533 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 7378670 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 8520812 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 28811426 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 15363113 | A-T | HOMO | INTRON | |
| SBS | Chr6 | 17047962 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 20085782 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 24660885 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 11481104 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g19400 |
| SBS | Chr7 | 17070887 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g29160 |
| SBS | Chr7 | 22368636 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 28128394 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 4485686 | G-A | HET | INTRON | |
| SBS | Chr7 | 7518758 | G-A | HET | INTRON | |
| SBS | Chr8 | 10894552 | A-G | HOMO | INTRON | |
| SBS | Chr8 | 24179869 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 8875671 | C-A | HOMO | INTRON |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 2956500 | 2956543 | 43 | |
| Deletion | Chr2 | 3884339 | 3884348 | 9 | |
| Deletion | Chr6 | 5390609 | 5390620 | 11 | |
| Deletion | Chr6 | 5485685 | 5485686 | 1 | |
| Deletion | Chr3 | 5744837 | 5744842 | 5 | |
| Deletion | Chr8 | 6196376 | 6196377 | 1 | |
| Deletion | Chr10 | 6257116 | 6257152 | 36 | |
| Deletion | Chr5 | 9384304 | 9384323 | 19 | |
| Deletion | Chr6 | 9480548 | 9480555 | 7 | |
| Deletion | Chr7 | 9847424 | 9847425 | 1 | |
| Deletion | Chr12 | 10834771 | 10834772 | 1 | |
| Deletion | Chr8 | 11637201 | 11637260 | 59 | |
| Deletion | Chr4 | 11800073 | 11800074 | 1 | |
| Deletion | Chr12 | 12046496 | 12046497 | 1 | |
| Deletion | Chr2 | 12185001 | 12413000 | 228000 | 30 |
| Deletion | Chr11 | 12187680 | 12187810 | 130 | |
| Deletion | Chr7 | 13226920 | 13226929 | 9 | LOC_Os07g23440 |
| Deletion | Chr3 | 15590401 | 15590421 | 20 | |
| Deletion | Chr10 | 15605986 | 15605989 | 3 | |
| Deletion | Chr1 | 15634355 | 15634359 | 4 | LOC_Os01g27960 |
| Deletion | Chr8 | 15722555 | 15722556 | 1 | |
| Deletion | Chr3 | 17151744 | 17151792 | 48 | |
| Deletion | Chr10 | 17582361 | 17582363 | 2 | |
| Deletion | Chr7 | 20355084 | 20355085 | 1 | |
| Deletion | Chr10 | 20611555 | 20611556 | 1 | |
| Deletion | Chr12 | 23057532 | 23057535 | 3 | |
| Deletion | Chr8 | 23075918 | 23075921 | 3 | |
| Deletion | Chr1 | 25823676 | 25823677 | 1 | |
| Deletion | Chr2 | 32104545 | 32104546 | 1 | |
| Deletion | Chr1 | 34886842 | 34886843 | 1 | |
| Deletion | Chr1 | 37759437 | 37759438 | 1 |
Insertions: 13
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 6848567 | 6849145 | |
| Inversion | Chr12 | 8418967 | 8419191 | |
| Inversion | Chr5 | 15705421 | 15705683 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 5944357 | Chr7 | 18282706 | |
| Translocation | Chr11 | 13893260 | Chr2 | 10875028 | |
| Translocation | Chr11 | 13893261 | Chr2 | 10875028 |
