Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN443-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN443-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 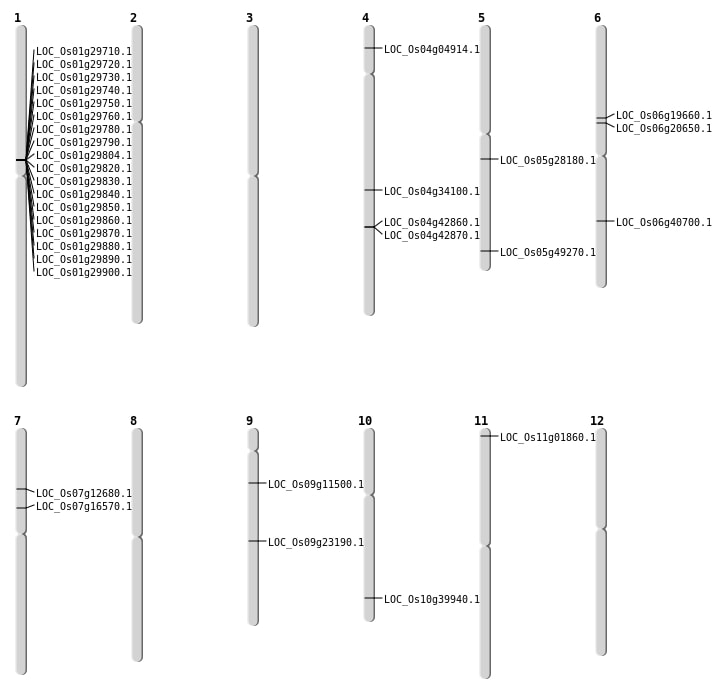
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17030644 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 17408895 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 19128884 | G-A | HET | INTRON | |
| SBS | Chr1 | 40418586 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 16129523 | A-G | HOMO | UTR_3_PRIME | |
| SBS | Chr11 | 456124 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g01860 |
| SBS | Chr12 | 24907969 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 23415461 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 17138315 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 24544751 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 15265270 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 18853762 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 19266921 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 20405083 | T-A | HET | INTRON | |
| SBS | Chr4 | 25369105 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g42860 |
| SBS | Chr4 | 25372721 | A-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 25372722 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g42870 |
| SBS | Chr5 | 14810957 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 15350868 | C-A | HET | INTRON | |
| SBS | Chr5 | 2399744 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 1474056 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 8629947 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 15735367 | G-C | HOMO | INTRON | |
| SBS | Chr7 | 9720622 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g16570 |
| SBS | Chr8 | 23075387 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 13749395 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g23190 |
| SBS | Chr9 | 15169825 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 3160507 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 6421488 | G-A | HET | STOP_GAINED | LOC_Os09g11500 |
| SBS | Chr9 | 7066170 | C-A | HET | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 52551 | 52552 | 1 | |
| Deletion | Chr5 | 187239 | 187241 | 2 | |
| Deletion | Chr7 | 1949198 | 1949220 | 22 | |
| Deletion | Chr4 | 2371509 | 2371510 | 1 | LOC_Os04g04914 |
| Deletion | Chr8 | 4110582 | 4110589 | 7 | |
| Deletion | Chr7 | 7244964 | 7244976 | 12 | LOC_Os07g12680 |
| Deletion | Chr6 | 11220663 | 11220682 | 19 | LOC_Os06g19660 |
| Deletion | Chr2 | 11566975 | 11566978 | 3 | |
| Deletion | Chr9 | 11743653 | 11743671 | 18 | |
| Deletion | Chr6 | 11899390 | 11899391 | 1 | LOC_Os06g20650 |
| Deletion | Chr7 | 13106324 | 13106334 | 10 | |
| Deletion | Chr3 | 15219614 | 15219615 | 1 | |
| Deletion | Chr5 | 16478640 | 16478642 | 2 | LOC_Os05g28180 |
| Deletion | Chr1 | 16649001 | 16763000 | 114000 | 18 |
| Deletion | Chr1 | 17023282 | 17023291 | 9 | |
| Deletion | Chr7 | 17031137 | 17031138 | 1 | |
| Deletion | Chr4 | 20653860 | 20653866 | 6 | LOC_Os04g34100 |
| Deletion | Chr10 | 21001457 | 21001459 | 2 | |
| Deletion | Chr8 | 21115621 | 21115624 | 3 | |
| Deletion | Chr11 | 21149409 | 21149410 | 1 | |
| Deletion | Chr5 | 21922412 | 21922438 | 26 | |
| Deletion | Chr6 | 24263236 | 24263238 | 2 | LOC_Os06g40700 |
| Deletion | Chr6 | 25874343 | 25874349 | 6 | |
| Deletion | Chr11 | 28177754 | 28177776 | 22 | |
| Deletion | Chr1 | 38637035 | 38637037 | 2 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr3 | 1248949 | 1248956 | 8 | |
| Insertion | Chr3 | 35802416 | 35802416 | 1 |
No Inversion
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 21388131 | Chr5 | 28265408 | LOC_Os05g49270 |
| Translocation | Chr10 | 21389198 | Chr5 | 28265405 | 2 |
| Translocation | Chr4 | 34971501 | Chr2 | 22517014 |
