Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4435-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4435-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 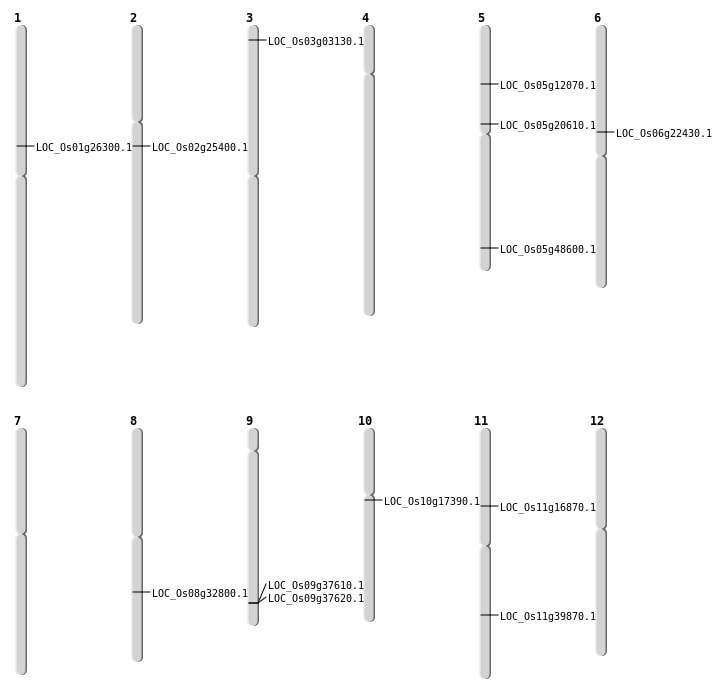
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10773039 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 14904229 | T-A | HOMO | STOP_GAINED | LOC_Os01g26300 |
| SBS | Chr1 | 23974703 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 24597979 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 27977696 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 30848799 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 32526199 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 34589623 | G-A | HET | INTRON | |
| SBS | Chr1 | 34959379 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 40397937 | G-T | HET | INTRON | |
| SBS | Chr11 | 14183016 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 17756024 | T-C | HOMO | INTRON | |
| SBS | Chr11 | 23767232 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g39870 |
| SBS | Chr11 | 9360104 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g16870 |
| SBS | Chr12 | 10797764 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 26002408 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr12 | 3868274 | A-T | HET | INTRON | |
| SBS | Chr2 | 1081875 | A-C | HET | INTRON | |
| SBS | Chr2 | 12174384 | A-C | HET | INTRON | |
| SBS | Chr2 | 14559110 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr2 | 14808992 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g25400 |
| SBS | Chr2 | 1687553 | T-A | HET | INTRON | |
| SBS | Chr3 | 18866506 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 12485316 | C-T | HET | INTRON | |
| SBS | Chr4 | 26616605 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 19690737 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 21803414 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 27007965 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 6904346 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 6904347 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g12070 |
| SBS | Chr6 | 16595957 | T-A | HOMO | INTRON | |
| SBS | Chr6 | 19960543 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr6 | 88804 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 18438055 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 28201369 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 10519098 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 14308206 | C-T | HOMO | INTRON | |
| SBS | Chr9 | 21455978 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 7881792 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 9168944 | C-T | HOMO | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 1071183 | 1071192 | 9 | |
| Deletion | Chr3 | 1321144 | 1321145 | 1 | LOC_Os03g03130 |
| Deletion | Chr3 | 1321871 | 1321873 | 2 | |
| Deletion | Chr10 | 1993496 | 1993503 | 7 | |
| Deletion | Chr12 | 2707747 | 2707753 | 6 | |
| Deletion | Chr10 | 6550087 | 6550088 | 1 | |
| Deletion | Chr2 | 7445310 | 7445312 | 2 | |
| Deletion | Chr12 | 8675058 | 8675066 | 8 | |
| Deletion | Chr4 | 8739129 | 8739131 | 2 | |
| Deletion | Chr10 | 8762863 | 8762864 | 1 | LOC_Os10g17390 |
| Deletion | Chr9 | 8823933 | 8823935 | 2 | |
| Deletion | Chr9 | 9735385 | 9735393 | 8 | |
| Deletion | Chr3 | 10169457 | 10169462 | 5 | |
| Deletion | Chr5 | 12092303 | 12092307 | 4 | LOC_Os05g20610 |
| Deletion | Chr1 | 12883923 | 12883925 | 2 | |
| Deletion | Chr6 | 13040447 | 13040471 | 24 | LOC_Os06g22430 |
| Deletion | Chr3 | 17109214 | 17109215 | 1 | |
| Deletion | Chr5 | 17341853 | 17341854 | 1 | |
| Deletion | Chr2 | 17801366 | 17801367 | 1 | |
| Deletion | Chr1 | 19052904 | 19052905 | 1 | |
| Deletion | Chr3 | 21070044 | 21070050 | 6 | |
| Deletion | Chr6 | 21829125 | 21829130 | 5 | |
| Deletion | Chr10 | 22440199 | 22440200 | 1 | |
| Deletion | Chr5 | 27863621 | 27863627 | 6 | LOC_Os05g48600 |
| Deletion | Chr4 | 29620042 | 29620043 | 1 |
Insertions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 18728645 | 18728645 | 1 | |
| Insertion | Chr11 | 18444205 | 18444205 | 1 | |
| Insertion | Chr11 | 9824913 | 9824913 | 1 | |
| Insertion | Chr12 | 22054647 | 22054647 | 1 | |
| Insertion | Chr12 | 3701952 | 3701985 | 34 | |
| Insertion | Chr2 | 11684888 | 11684911 | 24 | |
| Insertion | Chr3 | 14635456 | 14635511 | 56 | |
| Insertion | Chr3 | 5561828 | 5561845 | 18 | |
| Insertion | Chr4 | 26774142 | 26774142 | 1 | |
| Insertion | Chr4 | 29138315 | 29138365 | 51 | |
| Insertion | Chr5 | 12017251 | 12017271 | 21 | |
| Insertion | Chr6 | 20454098 | 20454098 | 1 | |
| Insertion | Chr7 | 3110897 | 3110897 | 1 | |
| Insertion | Chr8 | 20333407 | 20333407 | 1 | LOC_Os08g32800 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 4010415 | 4010492 | |
| Inversion | Chr12 | 7449852 | 7450116 | |
| Inversion | Chr9 | 21686027 | 21686191 | 2 |
| Inversion | Chr2 | 23078423 | 23078533 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 1800422 | Chr1 | 34373468 | |
| Translocation | Chr3 | 34304233 | Chr2 | 35453588 | |
| Translocation | Chr3 | 34304234 | Chr2 | 35432019 |
