Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4446-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4446-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 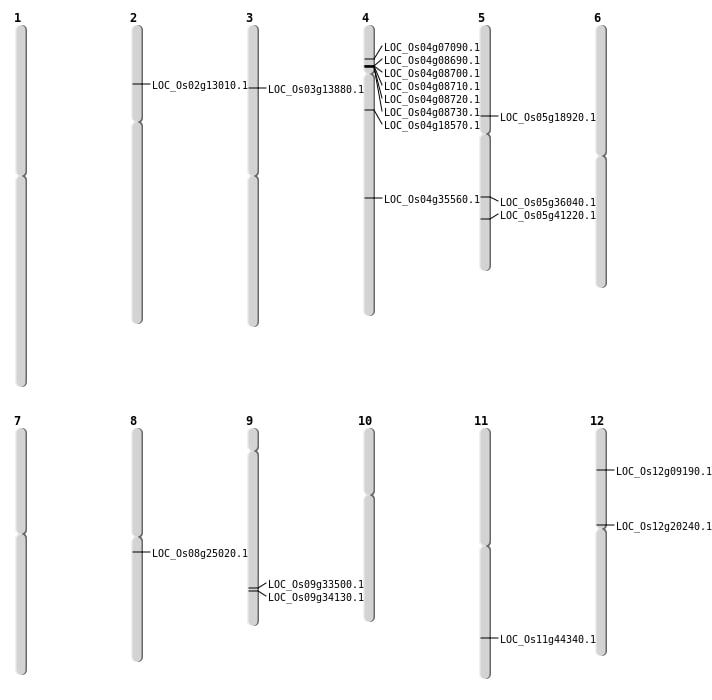
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 27803999 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 3998535 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 7253270 | G-T | HOMO | INTRON | |
| SBS | Chr10 | 10174613 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr10 | 224239 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 24955228 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 26801896 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g44340 |
| SBS | Chr11 | 9592985 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 11807953 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g20240 |
| SBS | Chr12 | 3017037 | A-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 24540359 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 28031153 | G-A | HET | INTRON | |
| SBS | Chr3 | 10691617 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 13937214 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 16841162 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 36225830 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 7709936 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 8908813 | G-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 10983196 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 10983197 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g18920 |
| SBS | Chr5 | 11091900 | T-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 2403371 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 21403442 | G-T | HOMO | UTR_5_PRIME | |
| SBS | Chr8 | 21403443 | A-T | HOMO | UTR_5_PRIME | |
| SBS | Chr8 | 21403444 | A-T | HOMO | UTR_5_PRIME | |
| SBS | Chr9 | 12620366 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 13392623 | T-C | HET | INTRON |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 629354 | 629355 | 1 | |
| Deletion | Chr11 | 1897182 | 1897184 | 2 | |
| Deletion | Chr12 | 2970452 | 2970453 | 1 | |
| Deletion | Chr4 | 3757023 | 3757024 | 1 | LOC_Os04g07090 |
| Deletion | Chr4 | 4718001 | 4736000 | 18000 | 5 |
| Deletion | Chr12 | 4797255 | 4797257 | 2 | LOC_Os12g09190 |
| Deletion | Chr4 | 5681837 | 5681843 | 6 | |
| Deletion | Chr2 | 6894596 | 6894597 | 1 | LOC_Os02g13010 |
| Deletion | Chr9 | 7424914 | 7424915 | 1 | |
| Deletion | Chr3 | 7529439 | 7529440 | 1 | LOC_Os03g13880 |
| Deletion | Chr8 | 7848616 | 7848617 | 1 | |
| Deletion | Chr4 | 10255009 | 10255016 | 7 | LOC_Os04g18570 |
| Deletion | Chr4 | 10444391 | 10444481 | 90 | |
| Deletion | Chr12 | 12625247 | 12625248 | 1 | |
| Deletion | Chr8 | 15189784 | 15189785 | 1 | LOC_Os08g25020 |
| Deletion | Chr8 | 16999177 | 16999178 | 1 | |
| Deletion | Chr5 | 17269199 | 17269200 | 1 | |
| Deletion | Chr10 | 18932212 | 18932216 | 4 | |
| Deletion | Chr6 | 20103193 | 20103198 | 5 | |
| Deletion | Chr7 | 20155953 | 20155968 | 15 | |
| Deletion | Chr11 | 20594436 | 20594479 | 43 | |
| Deletion | Chr1 | 21202648 | 21202649 | 1 | |
| Deletion | Chr4 | 21662901 | 21662912 | 11 | LOC_Os04g35560 |
| Deletion | Chr3 | 24139386 | 24139388 | 2 | |
| Deletion | Chr5 | 24140554 | 24140556 | 2 | LOC_Os05g41220 |
| Deletion | Chr8 | 24487079 | 24487091 | 12 | |
| Deletion | Chr12 | 25398084 | 25398116 | 32 | |
| Deletion | Chr3 | 27776916 | 27776920 | 4 | |
| Deletion | Chr2 | 28494364 | 28494437 | 73 | |
| Deletion | Chr1 | 28807960 | 28807969 | 9 | |
| Deletion | Chr1 | 31379690 | 31379691 | 1 |
Insertions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 8096577 | 8096587 | 11 | |
| Insertion | Chr11 | 11173699 | 11173699 | 1 | |
| Insertion | Chr12 | 19008062 | 19008067 | 6 | |
| Insertion | Chr3 | 10745204 | 10745204 | 1 | |
| Insertion | Chr4 | 25490476 | 25490476 | 1 | |
| Insertion | Chr4 | 32111892 | 32111892 | 1 | |
| Insertion | Chr5 | 21338291 | 21338291 | 1 | LOC_Os05g36040 |
| Insertion | Chr5 | 29932065 | 29932065 | 1 | |
| Insertion | Chr7 | 18061350 | 18061363 | 14 | |
| Insertion | Chr7 | 2834866 | 2834866 | 1 | |
| Insertion | Chr8 | 2285494 | 2285494 | 1 | |
| Insertion | Chr9 | 1827873 | 1827873 | 1 | |
| Insertion | Chr9 | 2067615 | 2067620 | 6 | |
| Insertion | Chr9 | 21123184 | 21123184 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 7254952 | 8197394 | |
| Inversion | Chr9 | 19752813 | 19876388 | LOC_Os09g33500 |
| Inversion | Chr9 | 19752842 | 19876401 | LOC_Os09g33500 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 20153712 | Chr6 | 7254943 | LOC_Os09g34130 |
| Translocation | Chr9 | 20154156 | Chr6 | 8197391 | LOC_Os09g34130 |
