Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN445-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN445-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 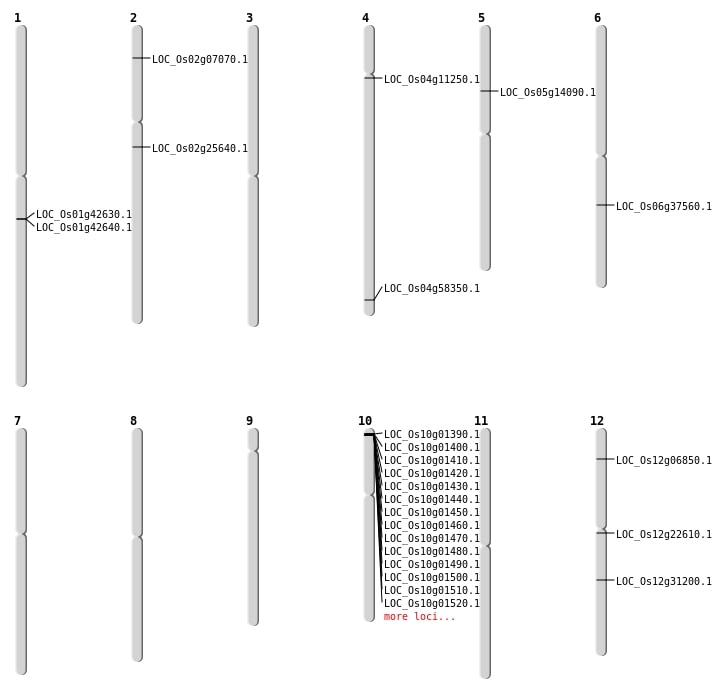
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18455408 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 27621839 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 27923323 | A-G | HET | INTRON | |
| SBS | Chr1 | 27923327 | C-G | HET | INTRON | |
| SBS | Chr1 | 32091044 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 3484864 | A-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 42966311 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 21693956 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr10 | 6155362 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 8605251 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 9638462 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 15984541 | G-A | HET | INTRON | |
| SBS | Chr11 | 27141597 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 12443694 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 16794221 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 14920701 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 3625887 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g07070 |
| SBS | Chr2 | 537357 | G-A | HET | INTRON | |
| SBS | Chr2 | 6870888 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 30014465 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 30850366 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 9494130 | C-T | HET | INTRON | |
| SBS | Chr3 | 9787118 | C-T | HET | INTRON | |
| SBS | Chr3 | 9787119 | A-T | HET | INTRON | |
| SBS | Chr4 | 34730367 | G-T | HET | STOP_GAINED | LOC_Os04g58350 |
| SBS | Chr4 | 5274695 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 16468477 | G-T | HET | UTR_3_PRIME | |
| SBS | Chr5 | 27209734 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 13137316 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 22246654 | G-A | HET | SPLICE_SITE_DONOR | LOC_Os06g37560 |
| SBS | Chr6 | 28171712 | T-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 11143768 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 17147624 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 3637930 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 11342069 | C-T | HET | INTRON | |
| SBS | Chr8 | 12231840 | T-C | HOMO | INTRON | |
| SBS | Chr8 | 1771439 | A-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 25244716 | T-G | HOMO | INTRON | |
| SBS | Chr8 | 7316041 | T-C | HOMO | INTRON | |
| SBS | Chr9 | 12334863 | C-T | HOMO | INTERGENIC |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 235001 | 414000 | 179000 | 23 |
| Deletion | Chr6 | 669543 | 669548 | 5 | |
| Deletion | Chr12 | 3325833 | 3325834 | 1 | LOC_Os12g06850 |
| Deletion | Chr8 | 7316044 | 7316052 | 8 | |
| Deletion | Chr11 | 7421020 | 7421022 | 2 | |
| Deletion | Chr5 | 7871444 | 7871453 | 9 | LOC_Os05g14090 |
| Deletion | Chr12 | 8302857 | 8302860 | 3 | |
| Deletion | Chr2 | 9771249 | 9771250 | 1 | |
| Deletion | Chr4 | 11466965 | 11467001 | 36 | |
| Deletion | Chr4 | 13040660 | 13040661 | 1 | |
| Deletion | Chr3 | 14496838 | 14496849 | 11 | |
| Deletion | Chr2 | 14992340 | 14992341 | 1 | LOC_Os02g25640 |
| Deletion | Chr11 | 15526571 | 15526583 | 12 | |
| Deletion | Chr11 | 19431565 | 19431566 | 1 | |
| Deletion | Chr1 | 24249288 | 24250500 | 1212 | 2 |
| Deletion | Chr11 | 26483199 | 26483249 | 50 | |
| Deletion | Chr1 | 27918522 | 27918523 | 1 | |
| Deletion | Chr6 | 28362244 | 28362245 | 1 | |
| Deletion | Chr2 | 31723151 | 31723152 | 1 | |
| Deletion | Chr4 | 34223729 | 34223738 | 9 |
Insertions: 6
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 12775849 | 13214338 | LOC_Os12g22610 |
| Inversion | Chr12 | 12776135 | 13214355 | LOC_Os12g22610 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 288999 | Chr8 | 20141139 | |
| Translocation | Chr10 | 9549409 | Chr4 | 6149050 | LOC_Os04g11250 |
| Translocation | Chr12 | 18786243 | Chr4 | 6148443 | LOC_Os12g31200 |
| Translocation | Chr12 | 18786246 | Chr10 | 9547461 | LOC_Os12g31200 |
| Translocation | Chr12 | 24854291 | Chr4 | 13170888 |
