Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4453-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4453-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 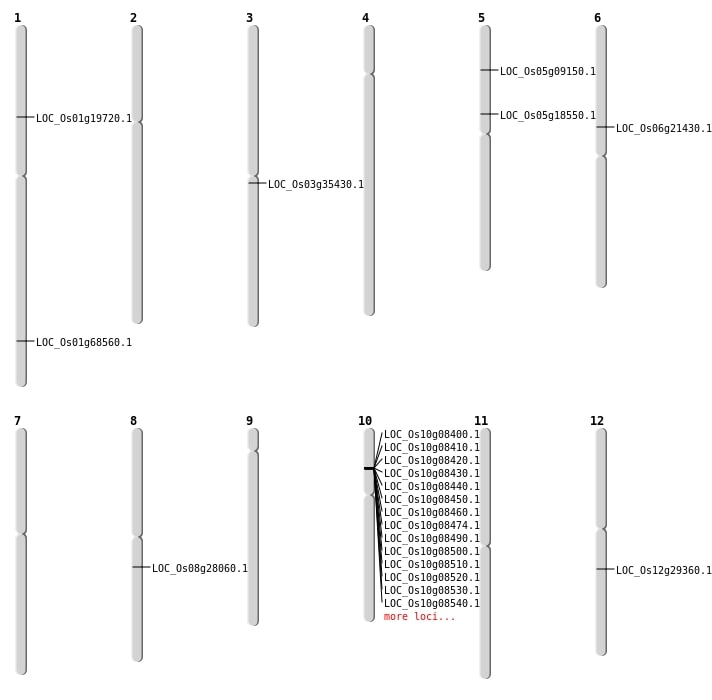
|
| Variant Information |
|---|
Single base substitutions: 28
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 26521878 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 39827462 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g68560 |
| SBS | Chr1 | 39827469 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g68560 |
| SBS | Chr1 | 40576115 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 12972495 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 12972501 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 7141857 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 7977296 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 3751589 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 7827967 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 19575665 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 19643953 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g35430 |
| SBS | Chr3 | 29247692 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 8177143 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 20833084 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 3039008 | C-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 8767479 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 10742132 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g18550 |
| SBS | Chr6 | 13018807 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 24803749 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 27607057 | A-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 41756 | C-T | HET | INTRON | |
| SBS | Chr7 | 11813127 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 16490501 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 20738785 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 20738786 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 16002905 | C-G | HOMO | SPLICE_SITE_REGION | |
| SBS | Chr9 | 2711574 | G-A | HET | SYNONYMOUS_CODING |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 898081 | 898085 | 4 | |
| Deletion | Chr2 | 2182909 | 2182910 | 1 | |
| Deletion | Chr4 | 3219442 | 3219443 | 1 | |
| Deletion | Chr9 | 3285419 | 3285420 | 1 | |
| Deletion | Chr10 | 4561001 | 4615000 | 54000 | 11 |
| Deletion | Chr10 | 4617001 | 4680000 | 63000 | 10 |
| Deletion | Chr8 | 5047933 | 5047934 | 1 | |
| Deletion | Chr5 | 5115132 | 5115136 | 4 | LOC_Os05g09150 |
| Deletion | Chr11 | 5919862 | 5919863 | 1 | |
| Deletion | Chr1 | 6194258 | 6194264 | 6 | |
| Deletion | Chr4 | 8633003 | 8633020 | 17 | |
| Deletion | Chr10 | 9383001 | 9684000 | 301000 | 52 |
| Deletion | Chr10 | 9461772 | 9461775 | 3 | |
| Deletion | Chr10 | 9685001 | 9732000 | 47000 | 9 |
| Deletion | Chr10 | 10055001 | 10137000 | 82000 | 12 |
| Deletion | Chr10 | 10143001 | 10189000 | 46000 | 8 |
| Deletion | Chr6 | 10866350 | 10866352 | 2 | |
| Deletion | Chr10 | 10921001 | 11150000 | 229000 | 30 |
| Deletion | Chr10 | 11152001 | 11438000 | 286000 | 41 |
| Deletion | Chr1 | 11184568 | 11184574 | 6 | LOC_Os01g19720 |
| Deletion | Chr6 | 12386172 | 12386173 | 1 | LOC_Os06g21430 |
| Deletion | Chr10 | 12388001 | 12439000 | 51000 | 10 |
| Deletion | Chr10 | 12446001 | 12643000 | 197000 | 32 |
| Deletion | Chr10 | 12644001 | 12675000 | 31000 | 4 |
| Deletion | Chr3 | 14093942 | 14093945 | 3 | |
| Deletion | Chr2 | 15862961 | 15862962 | 1 | |
| Deletion | Chr8 | 17106714 | 17106722 | 8 | LOC_Os08g28060 |
| Deletion | Chr12 | 17432328 | 17432329 | 1 | LOC_Os12g29360 |
| Deletion | Chr8 | 20272017 | 20272029 | 12 | |
| Deletion | Chr3 | 26058351 | 26058352 | 1 | |
| Deletion | Chr3 | 26762712 | 26762713 | 1 | |
| Deletion | Chr3 | 31329468 | 31329469 | 1 | |
| Deletion | Chr2 | 34476606 | 34476607 | 1 | |
| Deletion | Chr1 | 38616288 | 38616304 | 16 |
Insertions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 13438947 | 13438947 | 1 | |
| Insertion | Chr1 | 664612 | 664612 | 1 | |
| Insertion | Chr10 | 12628296 | 12628296 | 1 | LOC_Os10g24610 |
| Insertion | Chr10 | 6963763 | 6963765 | 3 | |
| Insertion | Chr11 | 1200977 | 1201003 | 27 | |
| Insertion | Chr12 | 14859883 | 14859883 | 1 | |
| Insertion | Chr2 | 26909876 | 26909877 | 2 | |
| Insertion | Chr2 | 31370667 | 31370667 | 1 | |
| Insertion | Chr3 | 20863896 | 20863896 | 1 | |
| Insertion | Chr3 | 34131742 | 34131742 | 1 | |
| Insertion | Chr4 | 2332596 | 2332596 | 1 | |
| Insertion | Chr4 | 2641367 | 2641368 | 2 | |
| Insertion | Chr4 | 4021831 | 4021831 | 1 | |
| Insertion | Chr5 | 17684059 | 17684059 | 1 | |
| Insertion | Chr5 | 4276329 | 4276329 | 1 | |
| Insertion | Chr6 | 18795365 | 18795366 | 2 | |
| Insertion | Chr6 | 7656560 | 7656567 | 8 | |
| Insertion | Chr7 | 9894037 | 9894040 | 4 | |
| Insertion | Chr9 | 13258549 | 13258549 | 1 | |
| Insertion | Chr9 | 15651874 | 15651912 | 39 | |
| Insertion | Chr9 | 7013095 | 7013095 | 1 |
No Inversion
No Translocation
