Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4468-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4468-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 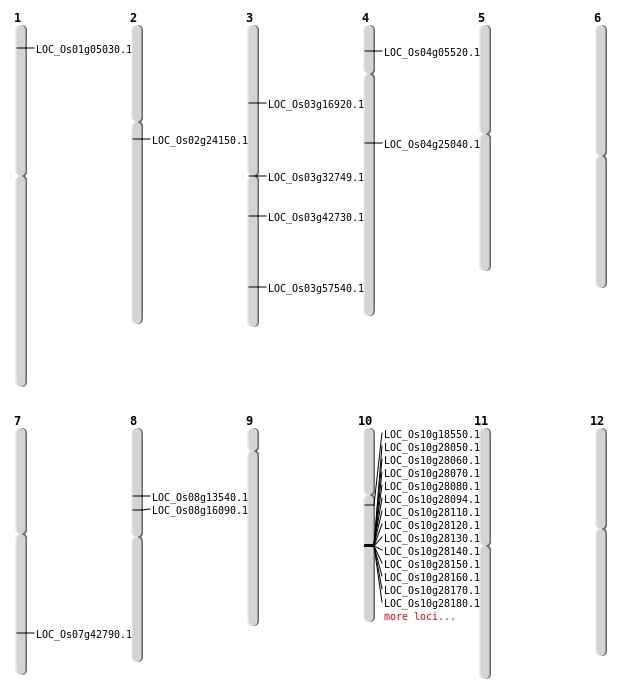
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13013520 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 23705337 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 28804874 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 14704117 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 17000809 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 1773349 | T-G | HET | INTERGENIC | |
| SBS | Chr10 | 18118254 | A-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 18513054 | T-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 20242493 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g37820 |
| SBS | Chr10 | 20242494 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g37820 |
| SBS | Chr10 | 8351801 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 1570331 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 18026136 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 3839290 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 15076212 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 18029678 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 13995289 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g24150 |
| SBS | Chr2 | 26638471 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 27710032 | C-G | HET | INTRON | |
| SBS | Chr2 | 6432302 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 14705017 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 32810389 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g57540 |
| SBS | Chr4 | 10694626 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 14439811 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g25040 |
| SBS | Chr4 | 23942283 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 2789748 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g05520 |
| SBS | Chr4 | 5376407 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 21049519 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 8278052 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 5997656 | A-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 14309500 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 26174917 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 7876656 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 20917881 | C-T | HET | INTRON | |
| SBS | Chr8 | 3278955 | T-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 8049933 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g13540 |
| SBS | Chr9 | 13672750 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 3575393 | A-T | HOMO | INTERGENIC |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 64012 | 64049 | 37 | |
| Deletion | Chr3 | 1503129 | 1503130 | 1 | |
| Deletion | Chr1 | 2367840 | 2367852 | 12 | LOC_Os01g05030 |
| Deletion | Chr2 | 5688233 | 5688239 | 6 | |
| Deletion | Chr8 | 7445002 | 7445003 | 1 | |
| Deletion | Chr10 | 13117777 | 13117779 | 2 | |
| Deletion | Chr3 | 13431058 | 13431063 | 5 | |
| Deletion | Chr10 | 14560001 | 14813000 | 253000 | 41 |
| Deletion | Chr2 | 14785311 | 14785312 | 1 | |
| Deletion | Chr10 | 14816001 | 14830000 | 14000 | 3 |
| Deletion | Chr10 | 14833001 | 14915000 | 82000 | 11 |
| Deletion | Chr7 | 16270766 | 16270767 | 1 | |
| Deletion | Chr5 | 17266396 | 17266397 | 1 | |
| Deletion | Chr8 | 19102461 | 19102466 | 5 | |
| Deletion | Chr1 | 19410311 | 19410312 | 1 | |
| Deletion | Chr4 | 20277370 | 20277374 | 4 | |
| Deletion | Chr5 | 21495805 | 21495813 | 8 | |
| Deletion | Chr6 | 23352365 | 23352457 | 92 | |
| Deletion | Chr8 | 24278937 | 24278938 | 1 | |
| Deletion | Chr4 | 34611416 | 34611418 | 2 | |
| Deletion | Chr1 | 38233800 | 38233801 | 1 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 25323770 | 25323770 | 1 | |
| Insertion | Chr11 | 3903761 | 3903761 | 1 | |
| Insertion | Chr2 | 6330548 | 6330548 | 1 | |
| Insertion | Chr4 | 1757511 | 1757511 | 1 | |
| Insertion | Chr4 | 2631922 | 2631922 | 1 | |
| Insertion | Chr5 | 20634220 | 20634221 | 2 | |
| Insertion | Chr6 | 23209053 | 23209053 | 1 | |
| Insertion | Chr8 | 9811693 | 9811693 | 1 | LOC_Os08g16090 |
| Insertion | Chr9 | 5650374 | 5650374 | 1 | |
| Insertion | Chr9 | 6881537 | 6881575 | 39 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 7483417 | 7815472 | |
| Inversion | Chr6 | 7787165 | 7815467 | |
| Inversion | Chr5 | 17337838 | 17337935 | |
| Inversion | Chr3 | 18746712 | 18750977 | LOC_Os03g32749 |
| Inversion | Chr3 | 18747106 | 18750984 | LOC_Os03g32749 |
| Inversion | Chr5 | 19301410 | 19510298 | |
| Inversion | Chr5 | 19301505 | 19510308 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 9411202 | Chr3 | 6132877 | LOC_Os10g18550 |
| Translocation | Chr10 | 9411736 | Chr3 | 6132903 | LOC_Os03g16920 |
| Translocation | Chr10 | 14482818 | Chr9 | 22293139 | |
| Translocation | Chr10 | 14485882 | Chr9 | 22293136 | |
| Translocation | Chr3 | 23790872 | Chr1 | 35655917 | LOC_Os03g42730 |
| Translocation | Chr3 | 23791474 | Chr1 | 35655917 | |
| Translocation | Chr7 | 25633898 | Chr5 | 22590045 | LOC_Os07g42790 |
