Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4475-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4475-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 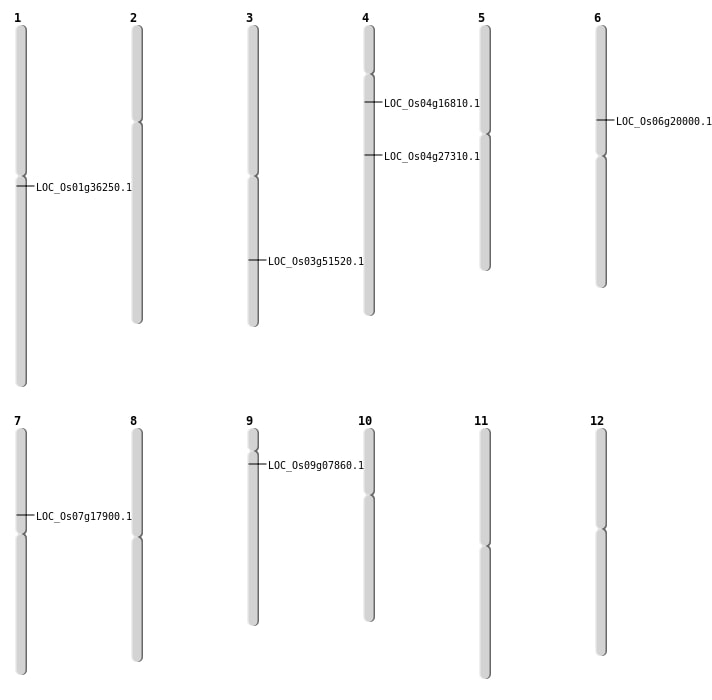
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20979412 | G-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 22665269 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 24292437 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 25540390 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 26299245 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 32175892 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 15259128 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 2146153 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 23141755 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 2400547 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 5888362 | C-T | HET | INTRON | |
| SBS | Chr11 | 18838613 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 28785949 | C-G | HET | UTR_3_PRIME | |
| SBS | Chr12 | 10261605 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 3012575 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 9024345 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 14609724 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 20984970 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 20984972 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 6223655 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 6623904 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 1491643 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 29476943 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g51520 |
| SBS | Chr3 | 30157618 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 6844473 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 7129959 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 16135742 | A-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g27310 |
| SBS | Chr4 | 22045500 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 9212804 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g16810 |
| SBS | Chr6 | 17635857 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 19961607 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr6 | 21776882 | T-G | HET | INTRON | |
| SBS | Chr7 | 12403177 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 13393652 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 21917741 | G-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 26844200 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 6070006 | T-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 8563563 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 395704 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 3997712 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g07860 |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 91931 | 91933 | 2 | |
| Deletion | Chr12 | 448471 | 448494 | 23 | |
| Deletion | Chr10 | 721041 | 721044 | 3 | |
| Deletion | Chr11 | 2523732 | 2523734 | 2 | |
| Deletion | Chr8 | 6369051 | 6369052 | 1 | |
| Deletion | Chr7 | 10592581 | 10592601 | 20 | LOC_Os07g17900 |
| Deletion | Chr6 | 11469668 | 11469670 | 2 | LOC_Os06g20000 |
| Deletion | Chr7 | 13566858 | 13566859 | 1 | |
| Deletion | Chr7 | 13579697 | 13579698 | 1 | |
| Deletion | Chr7 | 19581003 | 19581005 | 2 | |
| Deletion | Chr1 | 20010086 | 20010087 | 1 | |
| Deletion | Chr1 | 20070174 | 20070178 | 4 | LOC_Os01g36250 |
| Deletion | Chr2 | 22404805 | 22404806 | 1 | |
| Deletion | Chr1 | 25549775 | 25549776 | 1 | |
| Deletion | Chr5 | 29630550 | 29630557 | 7 | |
| Deletion | Chr5 | 29926506 | 29926509 | 3 | |
| Deletion | Chr2 | 33486149 | 33486157 | 8 | |
| Deletion | Chr4 | 33656234 | 33656256 | 22 | |
| Deletion | Chr1 | 34070691 | 34070692 | 1 | |
| Deletion | Chr1 | 34120460 | 34120461 | 1 |
Insertions: 9
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 12106082 | 12106259 |
No Translocation
