Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4476-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4476-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 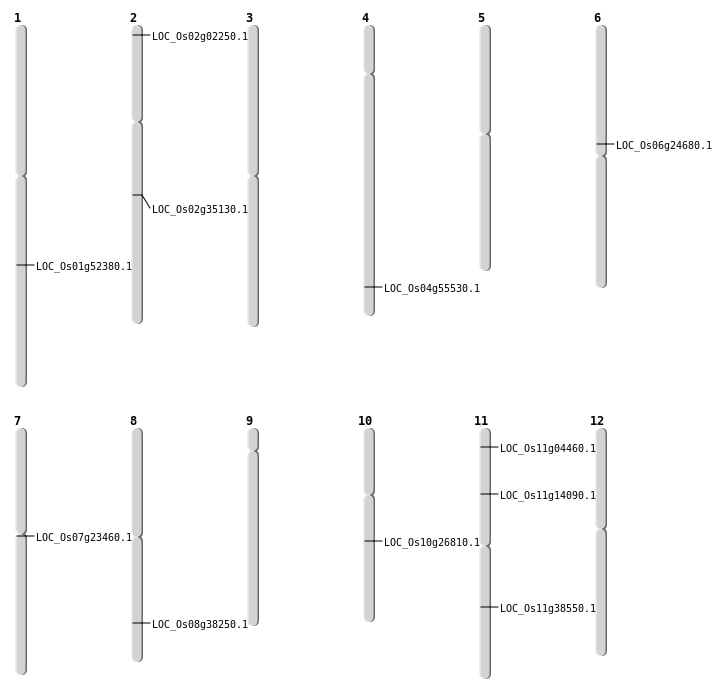
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14053701 | C-T | HOMO | INTRON | |
| SBS | Chr1 | 30091536 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g52380 |
| SBS | Chr1 | 34149247 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 12141132 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 14026546 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 14047583 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g26810 |
| SBS | Chr10 | 18124815 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 18368228 | C-G | HOMO | INTRON | |
| SBS | Chr11 | 5421629 | T-C | HET | INTRON | |
| SBS | Chr11 | 7848123 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g14090 |
| SBS | Chr12 | 15447934 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 15692222 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 20287861 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 8732490 | A-G | HET | INTRON | |
| SBS | Chr2 | 13533980 | A-G | HET | INTRON | |
| SBS | Chr2 | 15355536 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 21086010 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g35130 |
| SBS | Chr2 | 21939326 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 3552219 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 13901344 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 15524461 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 33236404 | T-C | HET | INTRON | |
| SBS | Chr4 | 2586215 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 28396797 | G-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 14585597 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 1976800 | A-G | HET | UTR_5_PRIME | |
| SBS | Chr5 | 2682378 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 11282564 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 12701406 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 14297973 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 22406834 | T-A | HOMO | INTRON | |
| SBS | Chr6 | 26169862 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 20866278 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 20866279 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 21716209 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 13968085 | T-C | HET | INTRON | |
| SBS | Chr8 | 19858098 | T-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 24245634 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g38250 |
| SBS | Chr9 | 10968900 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 20276932 | C-A | HET | INTERGENIC |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1861715 | 1861716 | 1 | LOC_Os11g04460 |
| Deletion | Chr10 | 2065656 | 2065663 | 7 | |
| Deletion | Chr7 | 2782215 | 2782248 | 33 | |
| Deletion | Chr3 | 2917220 | 2917222 | 2 | |
| Deletion | Chr4 | 4644455 | 4644456 | 1 | |
| Deletion | Chr7 | 5930674 | 5930681 | 7 | |
| Deletion | Chr8 | 5933067 | 5933068 | 1 | |
| Deletion | Chr7 | 7795237 | 7795238 | 1 | |
| Deletion | Chr8 | 8613585 | 8613586 | 1 | |
| Deletion | Chr10 | 9585164 | 9585165 | 1 | |
| Deletion | Chr5 | 10731606 | 10731607 | 1 | |
| Deletion | Chr7 | 11153010 | 11153011 | 1 | |
| Deletion | Chr8 | 13792219 | 13792279 | 60 | |
| Deletion | Chr7 | 13823875 | 13823884 | 9 | |
| Deletion | Chr8 | 13982746 | 13982747 | 1 | |
| Deletion | Chr6 | 14491248 | 14491249 | 1 | LOC_Os06g24680 |
| Deletion | Chr8 | 15070088 | 15070091 | 3 | |
| Deletion | Chr9 | 15890660 | 15890661 | 1 | |
| Deletion | Chr7 | 16123219 | 16123225 | 6 | |
| Deletion | Chr8 | 17319392 | 17319402 | 10 | |
| Deletion | Chr2 | 18678241 | 18678242 | 1 | |
| Deletion | Chr7 | 20237763 | 20237764 | 1 | |
| Deletion | Chr7 | 21204431 | 21204442 | 11 | |
| Deletion | Chr11 | 22843829 | 22843862 | 33 | LOC_Os11g38550 |
| Deletion | Chr3 | 23481158 | 23481192 | 34 | |
| Deletion | Chr8 | 24226414 | 24226427 | 13 | |
| Deletion | Chr8 | 25526633 | 25526634 | 1 | |
| Deletion | Chr1 | 25968020 | 25968039 | 19 | |
| Deletion | Chr5 | 27300827 | 27300872 | 45 | |
| Deletion | Chr3 | 28218034 | 28218128 | 94 | |
| Deletion | Chr1 | 30276538 | 30276539 | 1 |
Insertions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23745733 | 23745739 | 7 | |
| Insertion | Chr1 | 26959828 | 26959828 | 1 | |
| Insertion | Chr1 | 28925543 | 28925584 | 42 | |
| Insertion | Chr1 | 39284513 | 39284513 | 1 | |
| Insertion | Chr12 | 10988335 | 10988335 | 1 | |
| Insertion | Chr12 | 3839248 | 3839248 | 1 | |
| Insertion | Chr3 | 18716593 | 18716593 | 1 | |
| Insertion | Chr3 | 22759606 | 22759617 | 12 | |
| Insertion | Chr4 | 33034902 | 33034902 | 1 | LOC_Os04g55530 |
| Insertion | Chr4 | 5474629 | 5474629 | 1 | |
| Insertion | Chr7 | 16960128 | 16960128 | 1 | |
| Insertion | Chr7 | 4229277 | 4229277 | 1 | |
| Insertion | Chr8 | 17969694 | 17969742 | 49 | |
| Insertion | Chr8 | 24481449 | 24481469 | 21 | |
| Insertion | Chr8 | 8547068 | 8547068 | 1 | |
| Insertion | Chr8 | 95845 | 95848 | 4 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 733745 | 733855 | LOC_Os02g02250 |
| Inversion | Chr2 | 9592719 | 9708190 | |
| Inversion | Chr2 | 9708340 | 10175826 | |
| Inversion | Chr2 | 10174598 | 10175823 | |
| Inversion | Chr10 | 22202871 | 22203111 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 119173 | Chr2 | 3158072 | |
| Translocation | Chr7 | 119758 | Chr2 | 17979285 | |
| Translocation | Chr10 | 13248880 | Chr7 | 6861974 | LOC_Os07g23460 |
| Translocation | Chr5 | 27408695 | Chr4 | 28035492 |
